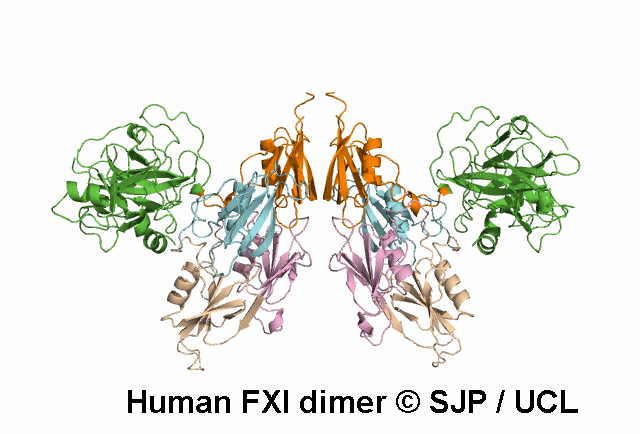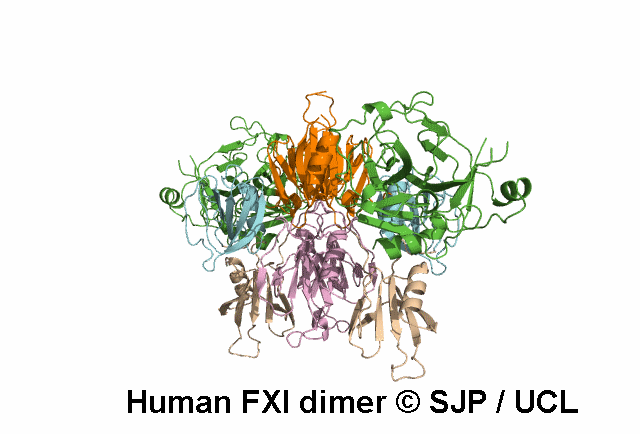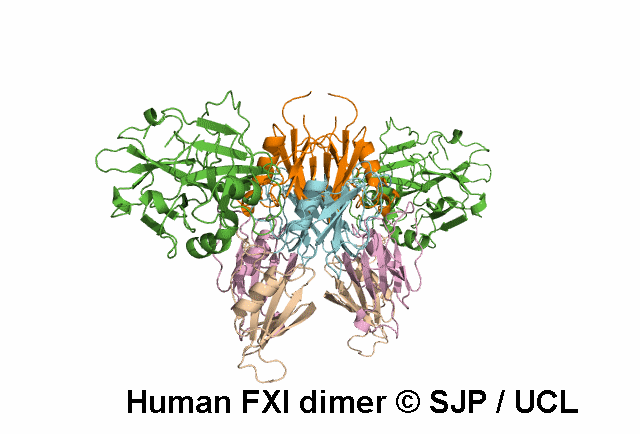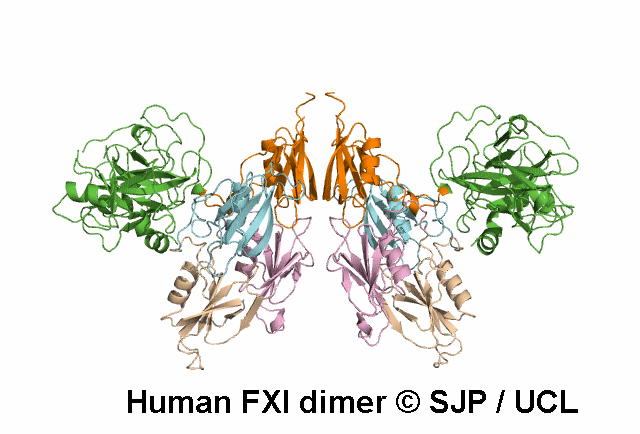In Depth Variant Analysis: c.688T>A (p.Cys230Ser)
Terms with a '*' next to them are explained on the Help Page .
c.688T>A
p.Cys230Ser (Legacy AA No. 212)
Variant Type:
Point
Domain:
Apple 3
Codon Change:
T>A
Variant Effect:
Missense
No. of Patients Reported:
4
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Cys230Ser in exon 7 suppresses a disulphide bond that maintains the 220-226 loop in the tertiary structure of the third apple domain. It is also possible that free cysteine affects other disulphide bonds such as the 226-255 bond. Aberrant or absent disulphide bonds may disrupt the protein structure leading to impaired secretion or function or increased clearance of misfolded proteins. Another mutation involving Cys230 has been described (Cys230Arg). The Cys230 residue is buried. As a consequence, the size or charge of the substituting Arg residue may contribute to protein structure disruption. In Cys230Ser however, neither size nor charge can account for the structure disruption, rather the suppression of disulphide bridges is likely to be the determinant of altered secretion. Quelin et al 2009Residue Information:
| Name | Type | Cyclic | Size | Position | Hydrophobicity | Charge | |
|---|---|---|---|---|---|---|---|
| |
|
|
|
|
|
|
|
| |
|
|
|
|
|
|
|
Substitution Analysis:
Structural Implications:
Cys230 is a buried residue (surface accessibility value = 0 ).
Cys230 is in a region of secondary structure within the FXI domains.
The DSSP assignment for this residue is ... H.
Cys230 is in a region of secondary structure within the FXI domains.
The DSSP assignment for this residue is ... H.
Hint: Left click mouse | To rotate the structure
Hint: Rotate mousewheel | To zoom in/out the structure
Hint: Right click mouse | to use applet control options
|
| Spacefill | |
| Cartoon | |
| Wireframe | |
| Trace | |
| Backbone | |
| Spin | |
| Background | |
| Disulphides | |
| Domains | |
| Alternative Colouring | |
| Labels |
Right Click on the molecule's screen for more options.
NOTE: If no Java applet appears or a Java error message is shown, please click the following LINK.