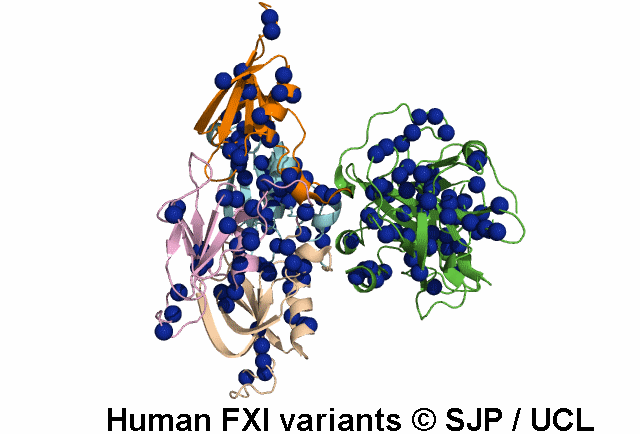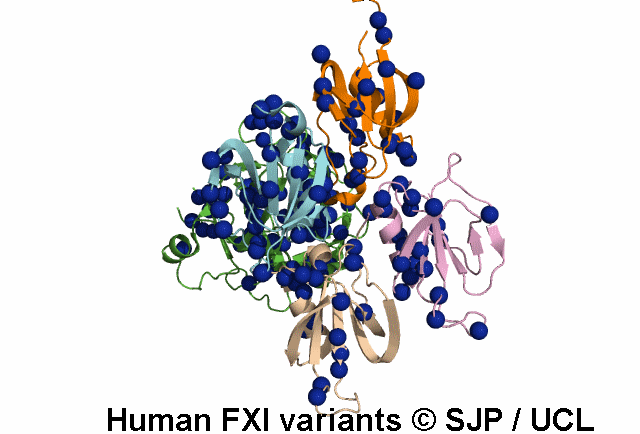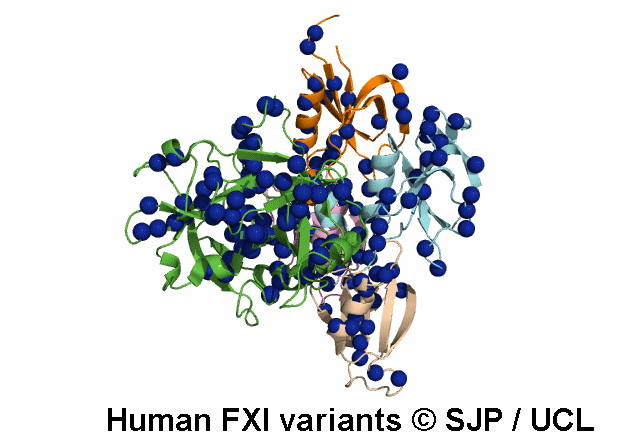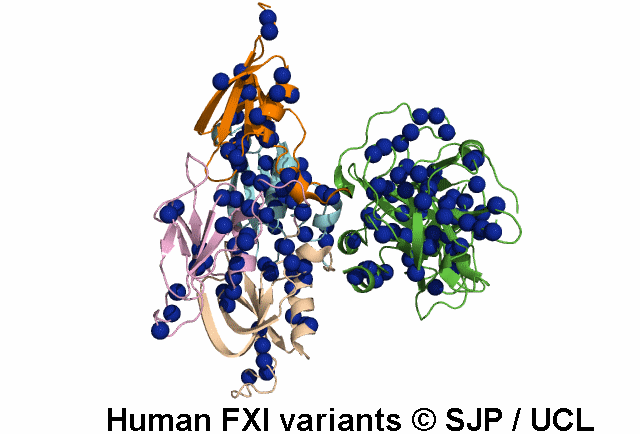
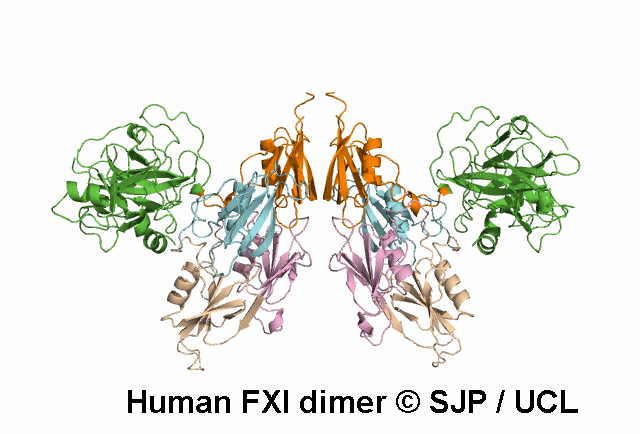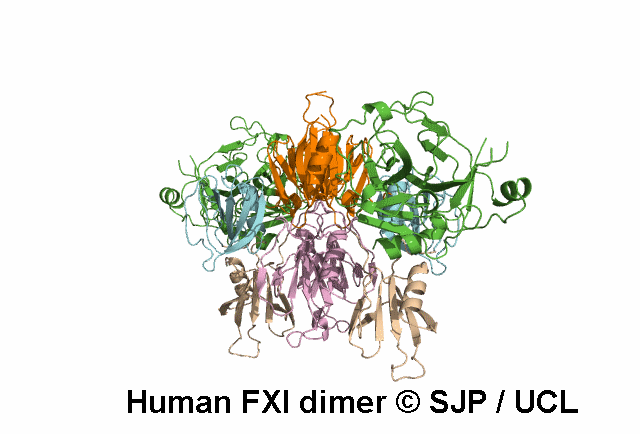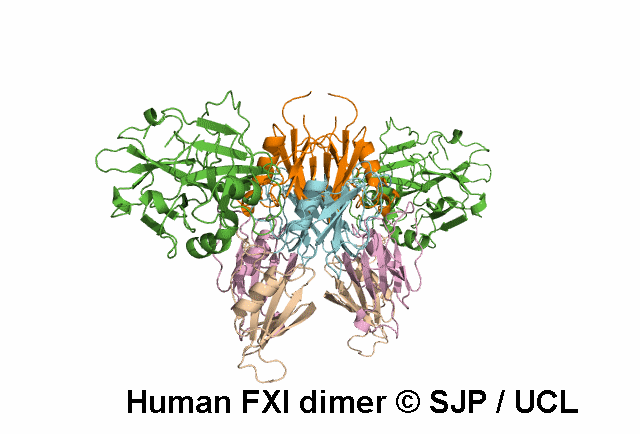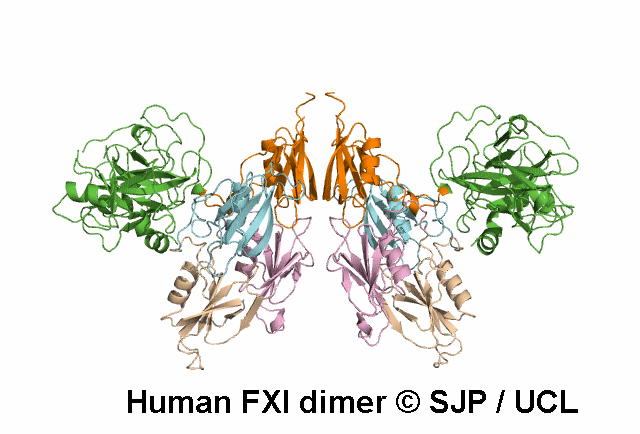In Depth Variant Analysis: c.1684G>A (p.Gly562Ser)
Terms with a '*' next to them are explained on the Help Page .
c.1684G>A
p.Gly562Ser (Legacy AA No. 544)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
G>A
Variant Effect:
Missense
No. of Patients Reported:
2
Phenotype:
U
Allele Count *:
10
Allele Number *:
251490
Allele Frequency *:
0.000040
Variant Comments & Reference:
Gly562Ser resides on a surface exposed loop within the SP domain and it is not clear whether this is the causative mutation because the patients mother has the mutation but has normal FXI:C levels. Saunders et al 2009Residue Information:
| Name | Type | Cyclic | Size | Position | Hydrophobicity | Charge | |
|---|---|---|---|---|---|---|---|
| |
|
|
|
|
|
|
|
| |
|
|
|
|
|
|
|
Substitution Analysis:
Structural Implications:
NOTE: If no Java applet appears or a Java error message is shown, please click the following LINK.