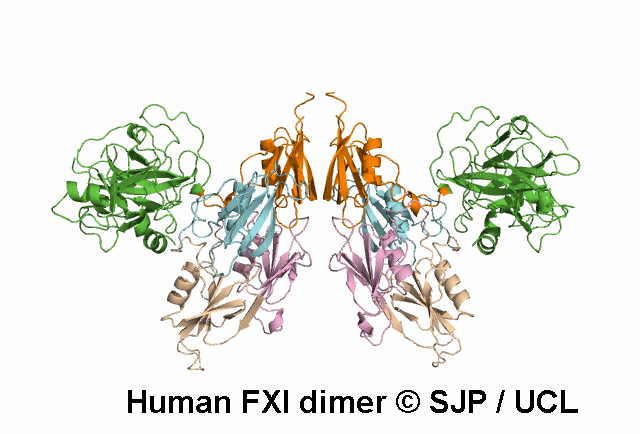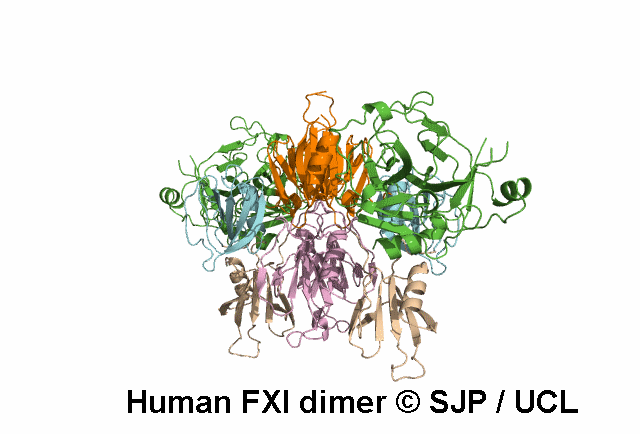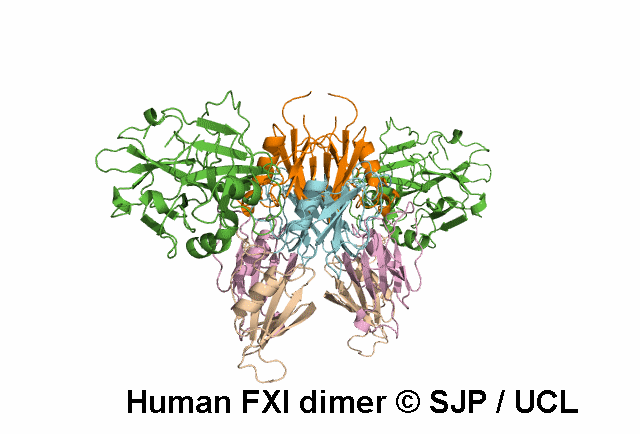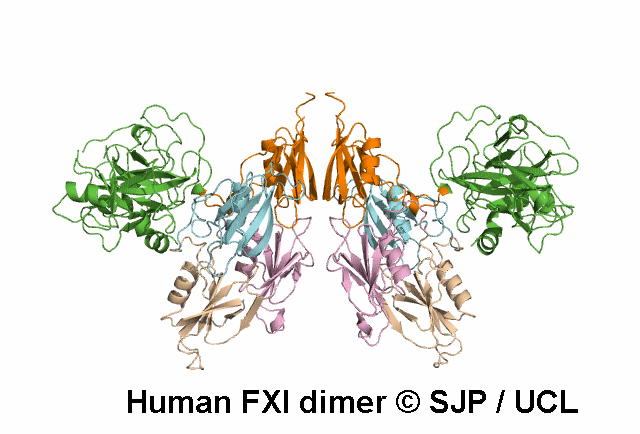In Depth Variant Analysis: c.1608G>C (p.Lys536Asn)
Terms with a '*' next to them are explained on the Help Page .
c.1608G>C
p.Lys536Asn (Legacy AA No. 518)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
G>C
Variant Effect:
Missense
No. of Patients Reported:
4
Phenotype:
I
Allele Count *:
1
Allele Number *:
251446
Allele Frequency *:
0.000004
Variant Comments & Reference:
The novel missense mutation FXI Lys536Asn leads to the replacement of the Lys536 which is part of a LxxxxxPxxxxxxC motif (x-variable amino acid) that occurs in several subfamilies of serine proteases. Although this motif is highly conserved between FXI and pre-kallikrein (86% sequence identity), the Lys536 is not conserved. On the other hand, this residue is conserved in human, murine and bovine FXI indicating that this is a FXI-specific amino acid. A molecular model of FXI predicts that the side-chain of this residue is on the surface of the molecule, possibly H-bonded to W515. Replacement of the positively charged lysine side chain by the smaller, neutral side chain of asparagine may interfere with the folding of the FXI molecule and/or secretion, leading to low antigen level. Alternatively, the secreted FXI variant may have a shortened plasma half-life. Ventura et al 2000, Esteban et al 2017Residue Information:
| Name | Type | Cyclic | Size | Position | Hydrophobicity | Charge | |
|---|---|---|---|---|---|---|---|
| |
|
|
|
|
|
|
|
| |
|
|
|
|
|
|
|
Substitution Analysis:
Structural Implications:
Lys536 is an exposed residue (surface accessibility value = 5 ).
Lys536 is in a region of secondary structure within the FXI domains.
The DSSP assignment for this residue is ... E.
Lys536 is in a region of secondary structure within the FXI domains.
The DSSP assignment for this residue is ... E.
Hint: Left click mouse | To rotate the structure
Hint: Rotate mousewheel | To zoom in/out the structure
Hint: Right click mouse | to use applet control options
|
| Spacefill | |
| Cartoon | |
| Wireframe | |
| Trace | |
| Backbone | |
| Spin | |
| Background | |
| Disulphides | |
| Domains | |
| Alternative Colouring | |
| Labels |
Right Click on the molecule's screen for more options.
NOTE: If no Java applet appears or a Java error message is shown, please click the following LINK.