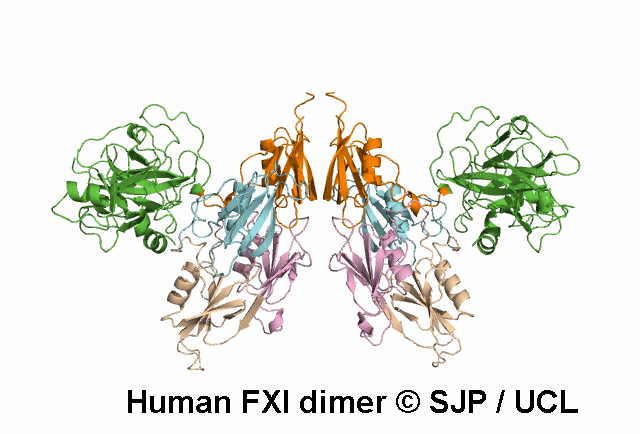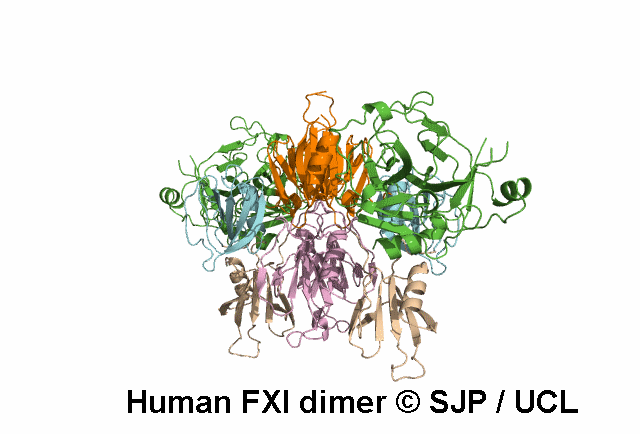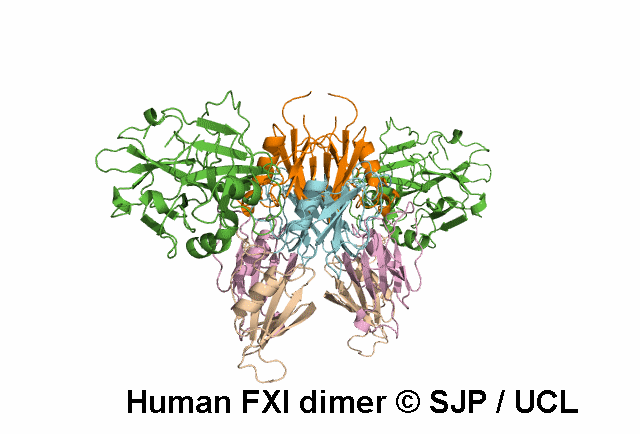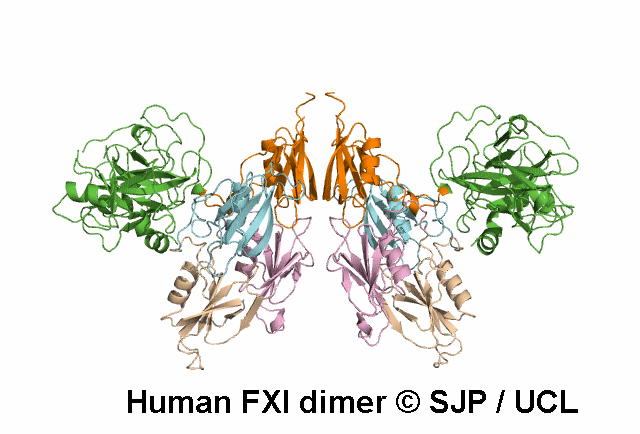In Depth Variant Analysis: c.1199C>T (p.Pro400Leu)
Terms with a '*' next to them are explained on the Help Page .
c.1199C>T
p.Pro400Leu (Legacy AA No. 382)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
C>T
Variant Effect:
Missense
No. of Patients Reported:
3
Phenotype:
I
Allele Count *:
2
Allele Number *:
251476
Allele Frequency *:
0.000008
Variant Comments & Reference:
Saunders et al 2009Residue Information:
| Name | Type | Cyclic | Size | Position | Hydrophobicity | Charge | |
|---|---|---|---|---|---|---|---|
| |
|
|
|
|
|
|
|
| |
|
|
|
|
|
|
|
Substitution Analysis:
Structural Implications:
Pro400 is a buried residue (surface accessibility value = 0 ).
Pro400 is in a random coil area of the FXI structure.
The DSSP assignment for this residue is ... T.
Pro400 is in a random coil area of the FXI structure.
The DSSP assignment for this residue is ... T.
Hint: Left click mouse | To rotate the structure
Hint: Rotate mousewheel | To zoom in/out the structure
Hint: Right click mouse | to use applet control options
|
| Spacefill | |
| Cartoon | |
| Wireframe | |
| Trace | |
| Backbone | |
| Spin | |
| Background | |
| Disulphides | |
| Domains | |
| Alternative Colouring | |
| Labels |
Right Click on the molecule's screen for more options.
NOTE: If no Java applet appears or a Java error message is shown, please click the following LINK.