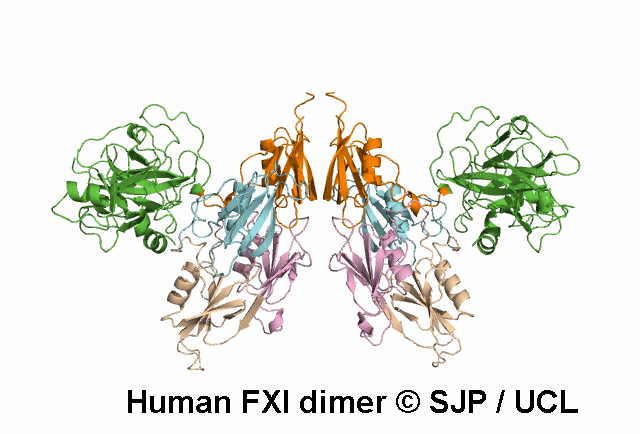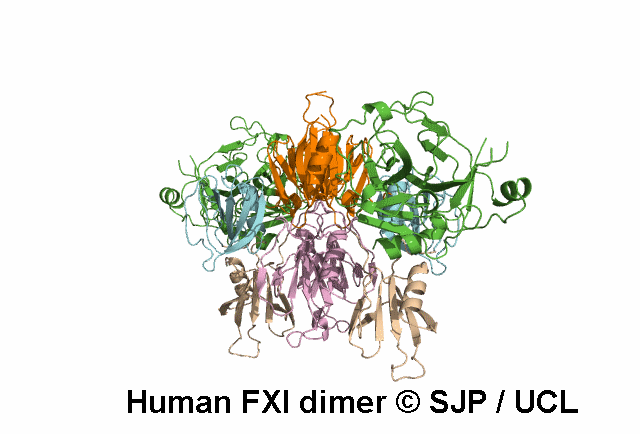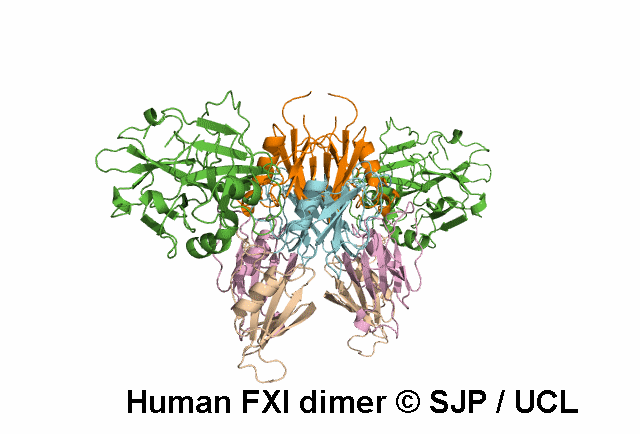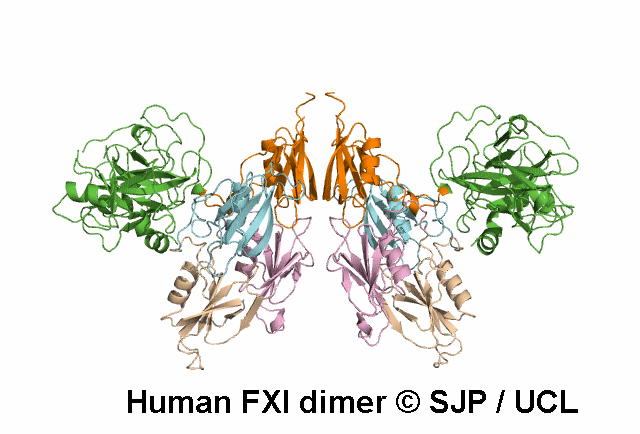In Depth Variant Analysis: c.1165G>A (p.Val389Ile)
Terms with a '*' next to them are explained on the Help Page .
c.1165G>A
p.Val389Ile (Legacy AA No. 371)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
G>A
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
II
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Residue Val389 is located one residue following the cleavage site of FXI (P2' position, according to the convention of numbering position around the scissile bond), thus is part of the activation loop. FXI antigen levels, measured in both conditioned media and lysates of cells expressing the mutant protein (in either the heterozygous or homozygous state), were not significantly different from those measured in wild-type samples. FXI specific activity was measured in conditioned media. The specific activity of the FXI Val389Ile protein was significantly reduced when compared with the wildtype one (30% and 80% for the heterozygous and the homozygous conditions, respectively). Given the proximity of Val389 to the FXI activation site, a possible interference on FXI activation was hypothesized and this mutation was found to be associated with a defect both in FXI activation (slower than normal), and in FIX activation (slightly delayed), thus supporting the role of residues neighboring the active site in influencing and stabilizing the enzyme active state. Bozzao et al 2007Residue Information:
| Name | Type | Cyclic | Size | Position | Hydrophobicity | Charge | |
|---|---|---|---|---|---|---|---|
| |
|
|
|
|
|
|
|
| |
|
|
|
|
|
|
|
Substitution Analysis:
Structural Implications:
Val389 is an exposed residue (surface accessibility value = 3 ).
Val389 is in a random coil area of the FXI structure.
The DSSP assignment for this residue is ... C.
Val389 is in a random coil area of the FXI structure.
The DSSP assignment for this residue is ... C.
Hint: Left click mouse | To rotate the structure
Hint: Rotate mousewheel | To zoom in/out the structure
Hint: Right click mouse | to use applet control options
|
| Spacefill | |
| Cartoon | |
| Wireframe | |
| Trace | |
| Backbone | |
| Spin | |
| Background | |
| Disulphides | |
| Domains | |
| Alternative Colouring | |
| Labels |
Right Click on the molecule's screen for more options.
NOTE: If no Java applet appears or a Java error message is shown, please click the following LINK.