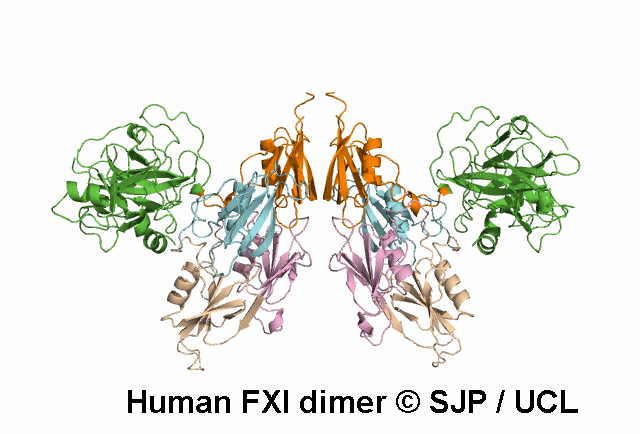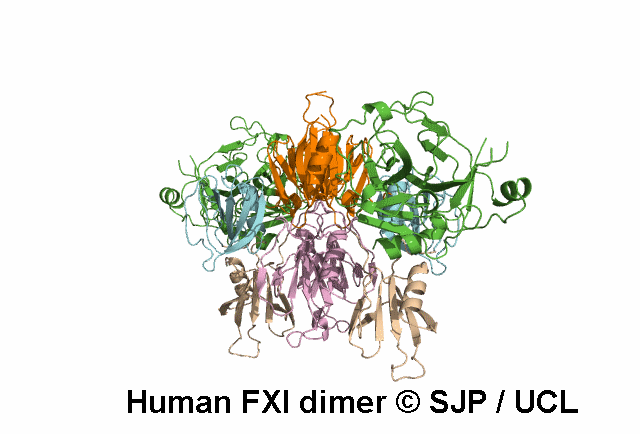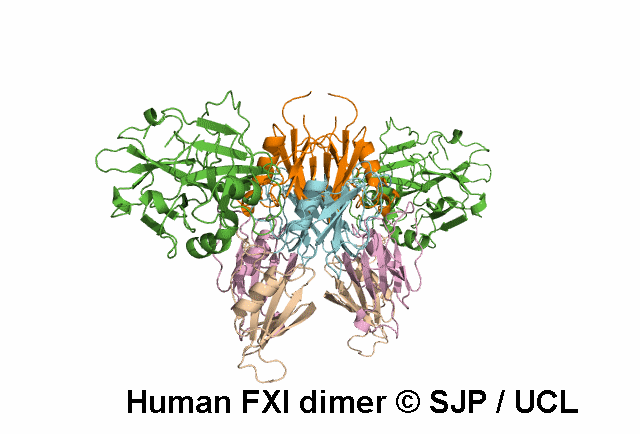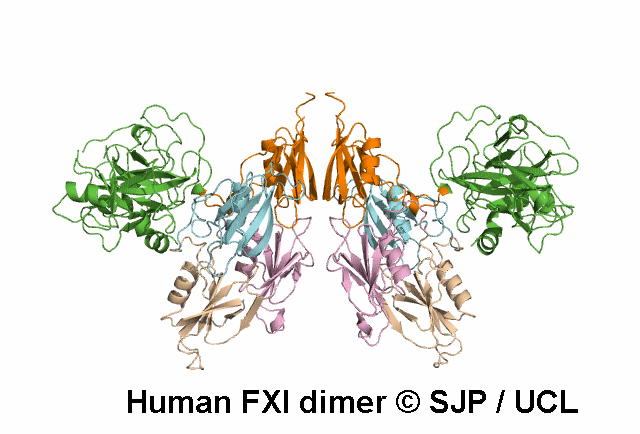Search Results: 3 unique variants retrieved
Terms with a '*' next to them are explained on the Help Page .
c.1102G>A
p.Gly368Arg (Legacy AA No. 350)
Variant Type:
Point
Domain:
Apple 4
Codon Change:
G>A
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
I
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
The Gly368Arg mutant is thought not to be secreted as the patient in which the mutation was found shows a parallel defect of both FXI:C and FXI:Ag levels. Quelin et al 2009 c.1103G>C
p.Gly368Ala (Legacy AA No. 350)
Variant Type:
Point
Domain:
Apple 4
Codon Change:
G>C
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
II
Allele Count *:
1
Allele Number *:
251366
Allele Frequency *:
0.000004
Variant Comments & Reference:
Decreases FXI dimerisation. Saunders et al 2009 c.1103G>A
p.Gly368Glu (Legacy AA No. 350)
Variant Type:
Point
Domain:
Apple 4
Codon Change:
G>A
Variant Effect:
Missense
No. of Patients Reported:
4
Phenotype:
I
Allele Count *:
3
Allele Number *:
251366
Allele Frequency *:
0.000012