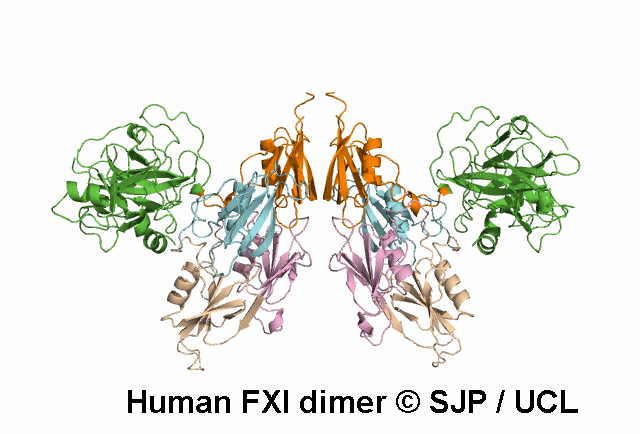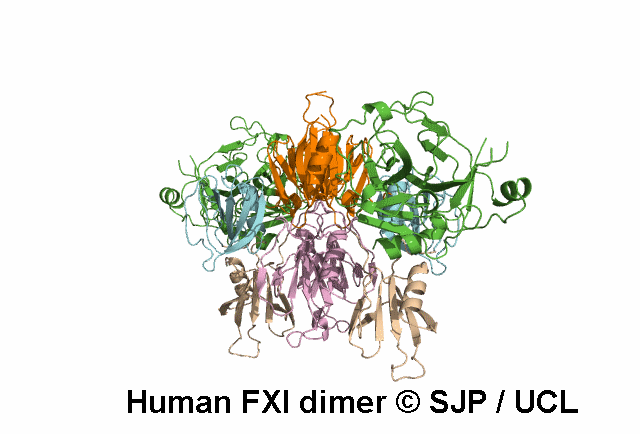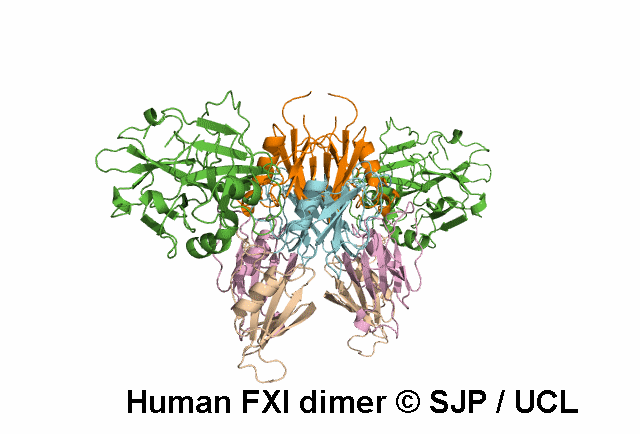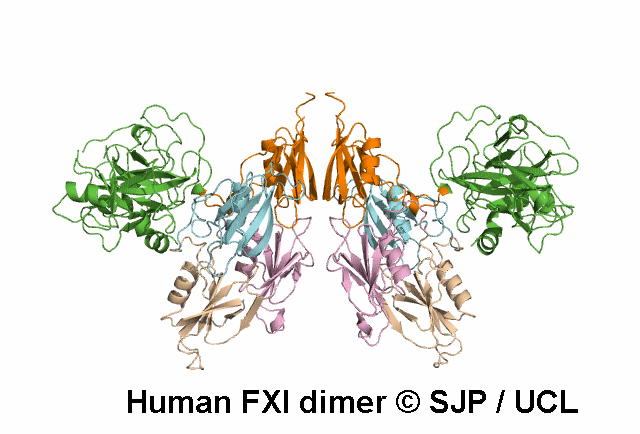Search Results: 3 unique variants retrieved
Terms with a '*' next to them are explained on the Help Page .
c.1106A>C
p.Tyr369Ser (Legacy AA No. 351)
Variant Type:
Point
Domain:
Apple 4
Codon Change:
A>C
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
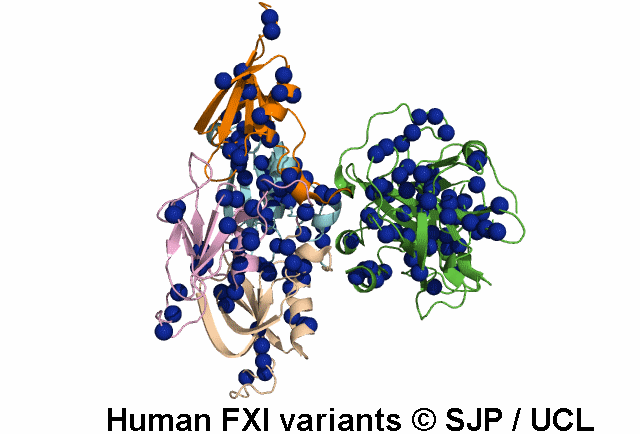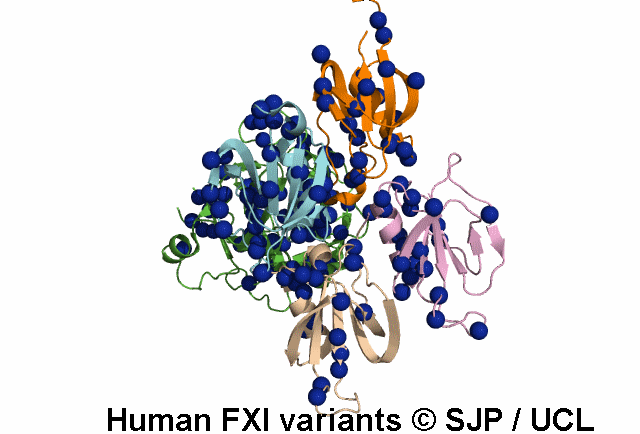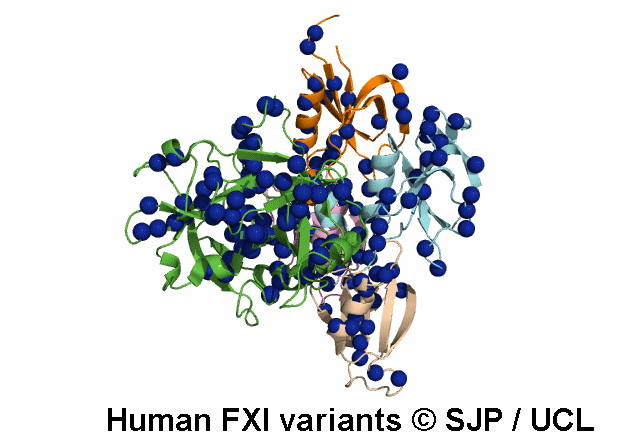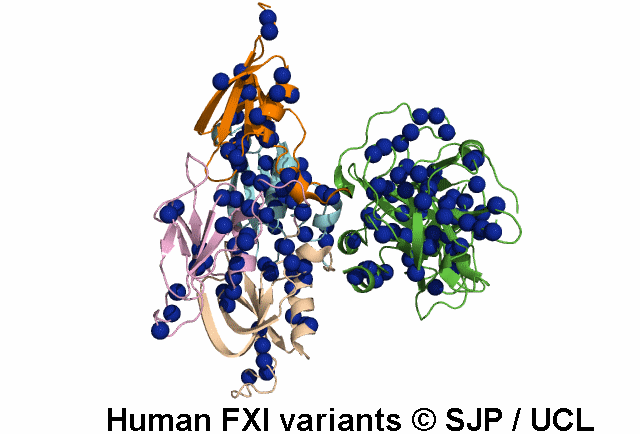
Tyr369 is highly conserved in the FXI amino acid sequences of related proteases of different species, indicating the importance of this residue to the structure of Ap4 domain in FXIa. Ap4 is important for the dimerisation of FXIa. A missense substitution at the adjoining residue Gly368Glu (factor XI Nagoya II) has been demonstrated (by expression studies) to have a profound effect on FXIa dimerization. It is possible that Tyr369Ser could have a similar effect on FXIa dimerisation. Kravtsov et al 2004, Jayandharan et al 2005 c.1107C>A
p.Tyr369* (Legacy AA No. 351)
Variant Type:
Point
Domain:
Apple 4
Codon Change:
C>A
Variant Effect:
Nonsense
No. of Patients Reported:
9
Phenotype:
U
Allele Count *:
10
Allele Number *:
282746
Allele Frequency *:
0.000035
Variant Comments & Reference:
Saunders et al 2005, Zhang et al 2020Structural Interpretation:
Structural analysis cannot be performed on this (Point | Nonsense) variant. c.1107C>T
p.Tyr369= (Legacy AA No. 351)
Variant Type:
Point
Domain:
Apple 4
Codon Change:
C>T
Variant Effect:
Silent
No. of Patients Reported:
3
Phenotype:
U
Allele Count *:
1
Allele Number *:
251352
Allele Frequency *:
0.000004