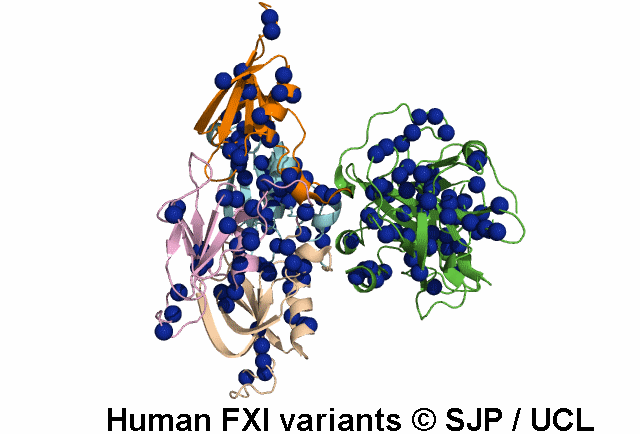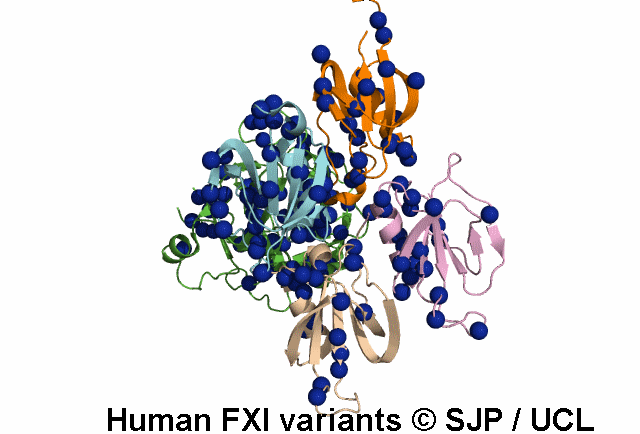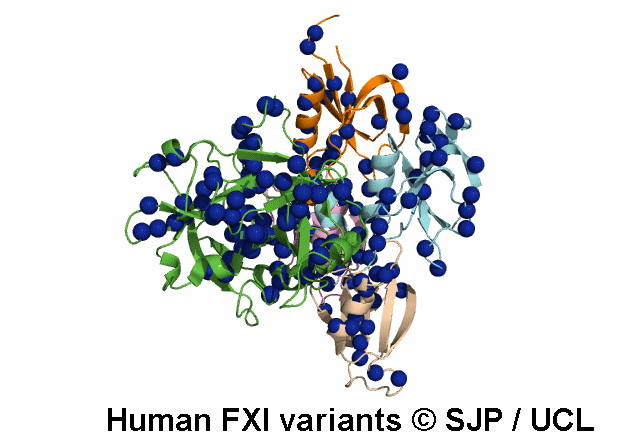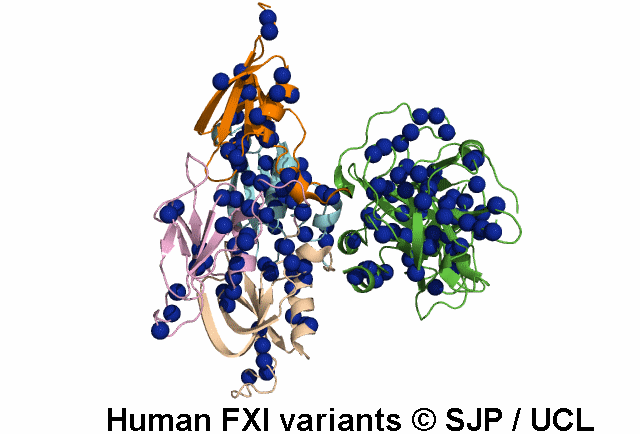
Search Results: 2 unique variants retrieved
Terms with a '*' next to them are explained on the Help Page .
c.151A>C
p.Thr51Pro (Legacy AA No. 33)
Variant Type:
Point
Domain:
Apple 1
Codon Change:
A>C
Variant Effect:
Missense
No. of Patients Reported:
2
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Thr51Pro and Thr51Ile. These changes convert hydrophilic and moderately sized threonine to hydrophobic, large proline and isoleucine, respectively. Thr51 lies in the first a-helix, in a buried region of Apple 1 domain and just next to a cysteine residue which forms the disulphide bond. On the other hand, the threonine residue is conserved in positions 51, 141, 231 and 322 of four Apple domains, in the same position relative to the a-helix of each domain. Therefore, it seems that this amino acid is essential for proper function of the protein and its alteration is not conservative. Fard-Esfahani et al 2008, Colakoglu et al 2018 c.152C>T
p.Thr51Ile (Legacy AA No. 33)
Variant Type:
Point
Domain:
Apple 1
Codon Change:
C>T
Variant Effect:
Missense
No. of Patients Reported:
0
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-