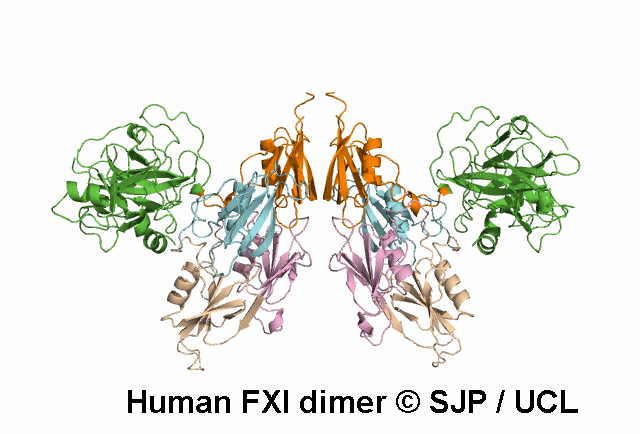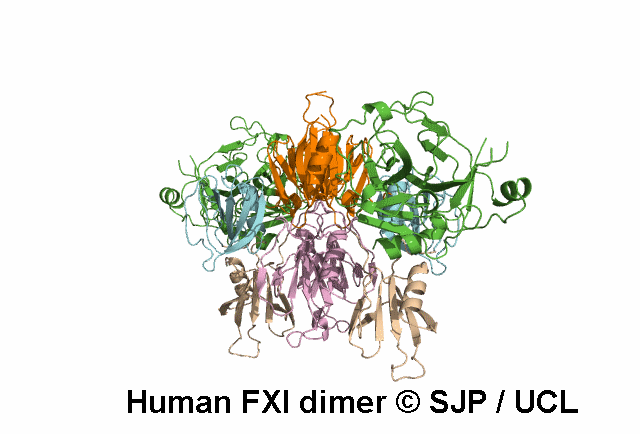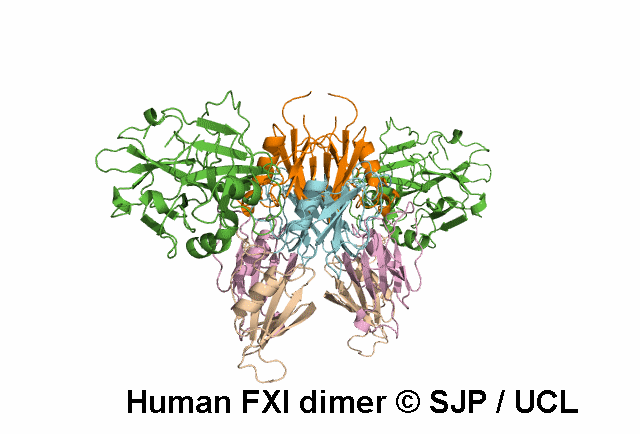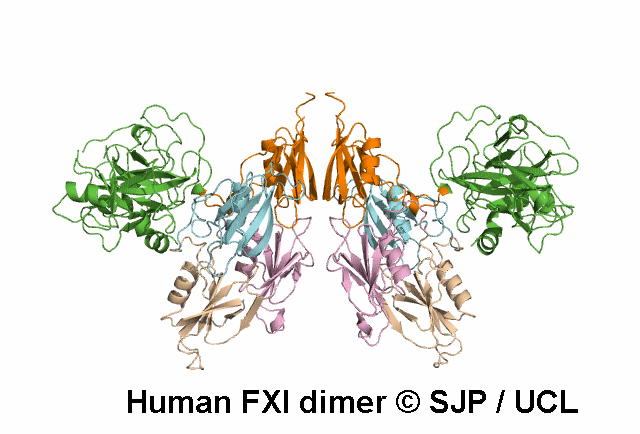In Depth Variant Analysis: c.359T>C (p.Met120Thr)
Terms with a '*' next to them are explained on the Help Page .
c.359T>C
p.Met120Thr (Legacy AA No. 102)
Variant Type:
Point
Domain:
Apple 2
Codon Change:
T>C
Variant Effect:
Missense
No. of Patients Reported:
2
Phenotype:
I
Allele Count *:
2
Allele Number *:
251262
Allele Frequency *:
0.000008
Variant Comments & Reference:
Met120 is a buried residue and is conserved across six species and prekallikrein. Replacement of the hydrophobic non-polar Met with a hydrophilic polar Thr is likely to be structurally disruptive. Expression studies indicate it is not expressed and type I. This amino acid substitution does not prevent the formation of dimers, despite the established role of the Ap2 domain in dimer formation. Mitchell et al 2006, Mitchell et al 2007Residue Information:
| Name | Type | Cyclic | Size | Position | Hydrophobicity | Charge | |
|---|---|---|---|---|---|---|---|
| |
|
|
|
|
|
|
|
| |
|
|
|
|
|
|
|
Substitution Analysis:
Structural Implications:
Met120 is a buried residue (surface accessibility value = 0 ).
Met120 is in a region of secondary structure within the FXI domains.
The DSSP assignment for this residue is ... E.
Met120 is in a region of secondary structure within the FXI domains.
The DSSP assignment for this residue is ... E.
Hint: Left click mouse | To rotate the structure
Hint: Rotate mousewheel | To zoom in/out the structure
Hint: Right click mouse | to use applet control options
|
| Spacefill | |
| Cartoon | |
| Wireframe | |
| Trace | |
| Backbone | |
| Spin | |
| Background | |
| Disulphides | |
| Domains | |
| Alternative Colouring | |
| Labels |
Right Click on the molecule's screen for more options.
NOTE: If no Java applet appears or a Java error message is shown, please click the following LINK.