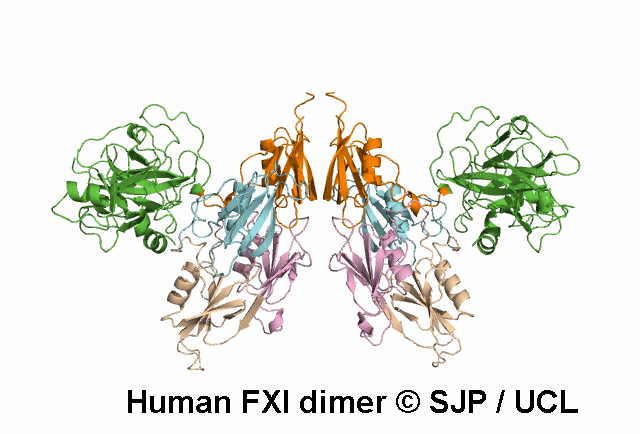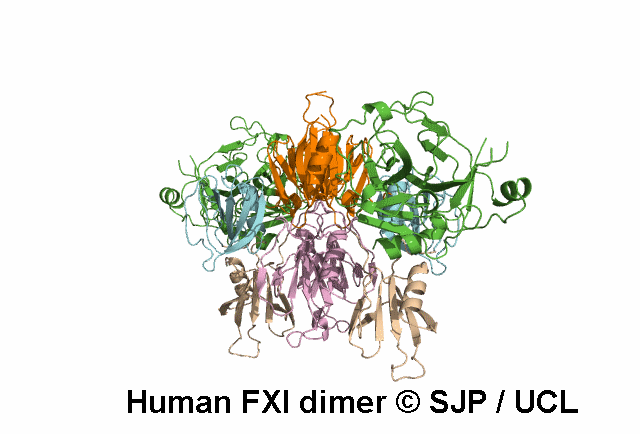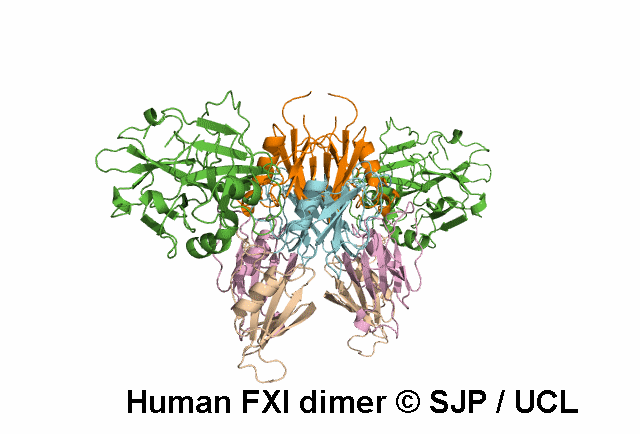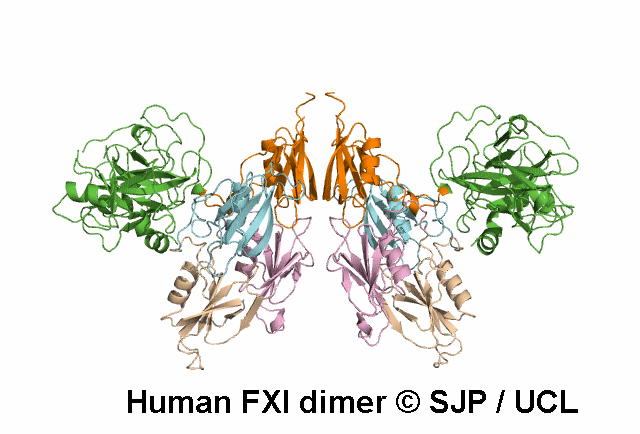Data Export Options:
Full List of Variants: 272 unique variants retrieved (displaying 50 entries per page). Scroll down to navigate to the next page(s).
Terms with a '*' next to them are explained on the Help Page .
c.764G>A
p.Cys255Tyr (Legacy AA No. 237)
Variant Type:
Point
Domain:
Apple 3
Codon Change:
G>A
Variant Effect:
Missense
No. of Patients Reported:
2
Phenotype:
I
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Expression studies showed that although mutant protein was synthesised in BHK transfected cells, it was not secreted. The medium of cells transfected with recombinant Cys255Tyr contained 3% of factor XI antigen secreted by normal cells but no demonstrable factor XI activity. The Cys255Tyr mutation disrupts the C226-C255 bond in Ap3. Zivelin et al 2002Structural Interpretation:
Please click HERE for in-depth variant analysis. c.783G>C
p.Glu261Asp (Legacy AA No. 243)
Variant Type:
Point
Domain:
Apple 3
Codon Change:
G>C
Variant Effect:
Missense
No. of Patients Reported:
2
Phenotype:
I
Allele Count *:
15
Allele Number *:
251328
Allele Frequency *:
0.000060
Variant Comments & Reference:
Saunders et al 2009Structural Interpretation:
Please click HERE for in-depth variant analysis. c.788G>A
p.Gly263Glu (Legacy AA No. 245)
Variant Type:
Point
Domain:
Apple 3
Codon Change:
G>A
Variant Effect:
Missense
No. of Patients Reported:
2
Phenotype:
I
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Hill et al 2005Structural Interpretation:
Please click HERE for in-depth variant analysis. c.797G>A
p.Ser266Asn (Legacy AA No. 248)
Variant Type:
Point
Domain:
Apple 3
Codon Change:
G>A
Variant Effect:
Missense
No. of Patients Reported:
2
Phenotype:
II
Allele Count *:
71
Allele Number *:
282734
Allele Frequency *:
0.000251
Variant Comments & Reference:
Mutant protein activates FXI normally in purified protein system and has normal activity, however activation of mutant protein by thrombin in presence of activated platelets is slower than normal. The mutation is associated with reduction in affinity for platelets. Sun et al 2001Structural Interpretation:
Please click HERE for in-depth variant analysis. c.801A>G
p.Thr267= (Legacy AA No. 249)
Variant Type:
Point
Domain:
Apple 3
Codon Change:
A>G
Variant Effect:
Silent
No. of Patients Reported:
0
Phenotype:
None
Allele Count *:
41844
Allele Number *:
282688
Allele Frequency *:
0.148022
Variant Comments & Reference:
Frequency in a random sample of normal volunteers is 19%, making this variant a polymorphism. Martincic et al 1998, Ventura et al 2000Structural Interpretation:
Structural analysis cannot be performed on this (Point | Silent) variant. c.802C>T
p.Arg268Cys (Legacy AA No. 250)
Variant Type:
Point
Domain:
Apple 3
Codon Change:
C>T
Variant Effect:
Missense
No. of Patients Reported:
5
Phenotype:
U
Allele Count *:
21
Allele Number *:
282758
Allele Frequency *:
0.000074
Variant Comments & Reference:
Bolton-Maggs et al 2003 (Abstract), Esteban et al 2017Structural Interpretation:
Please click HERE for in-depth variant analysis. c.803G>A
p.Arg268His (Legacy AA No. 250)
Variant Type:
Point
Domain:
Apple 3
Codon Change:
G>A
Variant Effect:
Missense
No. of Patients Reported:
2
Phenotype:
U
Allele Count *:
7
Allele Number *:
282734
Allele Frequency *:
0.000025
Variant Comments & Reference:
Homologue scanning mutagenesis studies of conformationally constrained synthetic peptides identified amino acid residues Arg268 (and Lys273) as being important for binding of FXI to platelets. Amino acid change at Arg268His is likely to cause disruption to normal platelet binding (but not as much as would Arg268Cys). Ho et al 2000, Duncan et al 2008Structural Interpretation:
Please click HERE for in-depth variant analysis. c.809A>T
p.Lys270Ile (Legacy AA No. 252)
Variant Type:
Point
Domain:
Apple 3
Codon Change:
A>T
Variant Effect:
Missense
No. of Patients Reported:
4
Phenotype:
I
Allele Count *:
148
Allele Number *:
282756
Allele Frequency *:
0.000523
Variant Comments & Reference:
Expression of mutant proteins in stably transfected cells showed a 73% reduction, but expression of the mutant protein in cell lysates was similar to normal - suggests mutant protein was synthesised but secretion was reduced. Dai et al 2004Structural Interpretation:
Please click HERE for in-depth variant analysis. c.829G>A
p.Gly277Ser (Legacy AA No. 259)
Variant Type:
Point
Domain:
Apple 3
Codon Change:
G>A
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
I
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Unpublished DataStructural Interpretation:
Please click HERE for in-depth variant analysis. c.841C>T
p.Gln281* (Legacy AA No. 263)
Variant Type:
Point
Domain:
Apple 3
Codon Change:
C>T
Variant Effect:
Nonsense
No. of Patients Reported:
22
Phenotype:
I
Allele Count *:
23
Allele Number *:
282772
Allele Frequency *:
0.000081
Variant Comments & Reference:
Saunders et al 2005, Shao et al 2016, Asselta et al 2017, Zhang et al 2020Structural Interpretation:
Structural analysis cannot be performed on this (Point | Nonsense) variant. c.861C>T
p.Ile287= (Legacy AA No. 269)
Variant Type:
Point
Domain:
Linker
Codon Change:
C>T
Variant Effect:
Silent
No. of Patients Reported:
0
Phenotype:
None
Allele Count *:
281
Allele Number *:
282734
Allele Frequency *:
0.000994
Variant Comments & Reference:
Polymorphism. Cargill et al 1999Structural Interpretation:
Structural analysis cannot be performed on this (Point | Silent) variant. c.865G>C
p.Val289Leu (Legacy AA No. 271)
Variant Type:
Point
Domain:
Apple 3
Codon Change:
G>C
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
1
Allele Number *:
251340
Allele Frequency *:
0.000004
Variant Comments & Reference:
As Val289 is the last codon of exon 8, located at its 3' splice donor junction, a mutation at this consensus splice sequence is predicted to abolish the physiological donor splice site for exon 8, resulting in an abnormal FXI transcript. Jayandharan et al 2005Structural Interpretation:
Please click HERE for in-depth variant analysis. c.901T>C
p.Phe301Leu (Legacy AA No. 283)
Variant Type:
Point
Domain:
Apple 4
Codon Change:
T>C
Variant Effect:
Missense
No. of Patients Reported:
22
Phenotype:
I
Allele Count *:
297
Allele Number *:
282824
Allele Frequency *:
0.001050
Variant Comments & Reference:
Jewish Type III. Mutant protein was secreted at reduced levels (8%) compared to wild type. Northern blot analysis demonstrated that wild type and mutant protein generated similar amounts of mRNA. Meijers et al 1992, Kravtsov et al 2004, Tiscia et al 2017Structural Interpretation:
Please click HERE for in-depth variant analysis. c.922A>T
p.Ile308Phe (Legacy AA No. 290)
Variant Type:
Point
Domain:
Apple 4
Codon Change:
A>T
Variant Effect:
Missense
No. of Patients Reported:
0
Phenotype:
None
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Polymorphism. Cargill et al 1999Structural Interpretation:
Please click HERE for in-depth variant analysis. c.923T>C
p.Ile308Thr (Legacy AA No. 290)
Variant Type:
Point
Domain:
Apple 4
Codon Change:
T>C
Variant Effect:
Missense
No. of Patients Reported:
0
Phenotype:
None
Allele Count *:
18
Allele Number *:
251388
Allele Frequency *:
0.000072
Variant Comments & Reference:
Polymorphism. Hill et al 2005Structural Interpretation:
Please click HERE for in-depth variant analysis. c.938G>T
p.Ser313Ile (Legacy AA No. 295)
Variant Type:
Point
Domain:
Apple 4
Codon Change:
G>T
Variant Effect:
Missense
No. of Patients Reported:
8
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Variant predicted to disrupt hydrogen-bond formation, altering protein structure and function. Wang et al 2019, Xia et al 2020Structural Interpretation:
Please click HERE for in-depth variant analysis. c.943G>A
p.Glu315Lys (Legacy AA No. 297)
Variant Type:
Point
Domain:
Apple 4
Codon Change:
G>A
Variant Effect:
Missense
No. of Patients Reported:
11
Phenotype:
I
Allele Count *:
8
Allele Number *:
282756
Allele Frequency *:
0.000028
Variant Comments & Reference:
Expression of Glu315Lys in BHK cells revealed impaired secretion (4.5% of wild-type FXI) despite the presence of 70% of wild-type FXI antigen in cell lysate, which was shown to be a dimer by western blot analysis. Zucker et al 2007, Tiscia et al 2017Structural Interpretation:
Please click HERE for in-depth variant analysis. c.943G>T
p.Glu315* (Legacy AA No. 297)
Variant Type:
Point
Domain:
Apple 4
Codon Change:
G>T
Variant Effect:
Nonsense
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Hopmeier et al 2004Structural Interpretation:
Structural analysis cannot be performed on this (Point | Nonsense) variant. c.959T>C
p.Leu320Pro (Legacy AA No. 302)
Variant Type:
Point
Domain:
Apple 4
Codon Change:
T>C
Variant Effect:
Missense
No. of Patients Reported:
0
Phenotype:
I
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Amount of mutant protein secreted from cells in vitro is reduced. Leu320 occurs adjacent to Cys321, which forms a disulfide bond with Cys327. Structural predictions indicate that the substitution of Pro for Leu320 causes the helical conformation at Cys317, Gln318, and Lys319 of the normal protein to be changed to turn and coil conformations. These changes in protein conformation might prevent the formation of the Cys321- Cys327 disulfide bond, altering the structure of the fourth apple domain. Pugh et al 1995Structural Interpretation:
Please click HERE for in-depth variant analysis. c.965C>T
p.Thr322Ile (Legacy AA No. 304)
Variant Type:
Point
Domain:
Apple 4
Codon Change:
C>T
Variant Effect:
Missense
No. of Patients Reported:
3
Phenotype:
I
Allele Count *:
4
Allele Number *:
251374
Allele Frequency *:
0.000016
Variant Comments & Reference:
Thr322Ile was introduced into factor XI cDNA by site directed mutagenesis and cloned into pEXV-3. Amount of mutant protein secreted from cells in vitro is reduced. Pugh et al 1995Structural Interpretation:
Please click HERE for in-depth variant analysis. c.973G>T
p.Val325Phe (Legacy AA No. 307)
Variant Type:
Point
Domain:
Apple 4
Codon Change:
G>T
Variant Effect:
Missense
No. of Patients Reported:
3
Phenotype:
U
Allele Count *:
2
Allele Number *:
251386
Allele Frequency *:
0.000008
Variant Comments & Reference:
Val325Phe is located buried at the surface of the Ap4 domain, and lies at the interface between the Ap1 and Ap4 domains, thus this mutation may disrupt the packing between the Ap domains. Saunders et al 2009Structural Interpretation:
Please click HERE for in-depth variant analysis. Unspecified (R326H)
p.Arg326His (Legacy AA No. 308)
Variant Type:
Point
Domain:
Apple 4
Codon Change:
Variant Effect:
Missense
No. of Patients Reported:
0
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Shao et al 2016Structural Interpretation:
Please click HERE for in-depth variant analysis. c.976C>T
p.Arg326Cys (Legacy AA No. 308)
Variant Type:
Point
Domain:
Apple 4
Codon Change:
C>T
Variant Effect:
Missense
No. of Patients Reported:
4
Phenotype:
I
Allele Count *:
6
Allele Number *:
251384
Allele Frequency *:
0.000024
Variant Comments & Reference:
Introduction of a Cys residue may disrupt disulphide bridge formation. Arg326 lies close to the part of the Ap4 domain responsible for extensive non-covalent interactions between the FXI monomers. Saunders et al 2009Structural Interpretation:
Please click HERE for in-depth variant analysis. c.981C>A
p.Cys327* (Legacy AA No. 309)
Variant Type:
Point
Domain:
Apple 4
Codon Change:
C>A
Variant Effect:
Nonsense
No. of Patients Reported:
1
Phenotype:
I
Allele Count *:
1
Allele Number *:
251406
Allele Frequency *:
0.000004
Variant Comments & Reference:
Cys327* occurs in the Ap4 domain, with disruption of the Cys321-Cys327 pairing in the inner loop. Castaman et al 2008Structural Interpretation:
Structural analysis cannot be performed on this (Point | Nonsense) variant. c.992C>T
p.Thr331Ile (Legacy AA No. 313)
Variant Type:
Point
Domain:
Apple 4
Codon Change:
C>T
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Interestingly, this mutation is similar to the Thr51Ile conversion but occurs in Ap4. This Thr residue, Thr331, which is laid in the extended strand positioned after alpha-helix, is conserved in positions 60, 150, 240 and 331 of Apple 1, 2, 3 and 4 domains, respectively. The major role of Apple 4 is to mediate dimer formation of two identical polypeptide chains and the Thr331Ile change may distort the conformation of the Apple 4 domain and interfere with its function. Fard-Esfahani et al 2008Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1016G>T
p.Cys339Phe (Legacy AA No. 321)
Variant Type:
Point
Domain:
Apple 4
Codon Change:
G>T
Variant Effect:
Missense
No. of Patients Reported:
0
Phenotype:
None
Allele Count *:
566
Allele Number *:
282858
Allele Frequency *:
0.002001
Variant Comments & Reference:
Polymorphism. Cys339 pairs with other Cys339 of Ap4 at dimerisation - however it is not required for dimerisation. SDS-PAGE analysis revealed only monomer of FXI present, but mutant protein expressed in BHK cells and normal FXI secretion and activity was present. Meijers 1992, Zivelin et al 2002Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1021G>A
p.Glu341Lys (Legacy AA No. 323)
Variant Type:
Point
Domain:
Apple 4
Codon Change:
G>A
Variant Effect:
Missense
No. of Patients Reported:
6
Phenotype:
I
Allele Count *:
4
Allele Number *:
251454
Allele Frequency *:
0.000016
Variant Comments & Reference:
Glu341 lies close to the part of the Ap4 domain responsible for extensive non-covalent interactions between the FXI monomers. Amount of mutant protein secreted from cells in vitro is reduced. Pugh et al 1995, Saunders et al 2009, Zhang et al 2020Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1026G>T
p.Gly342= (Legacy AA No. 324)
Variant Type:
Point
Domain:
Apple 4
Codon Change:
G>T
Variant Effect:
Silent
No. of Patients Reported:
3
Phenotype:
I
Allele Count *:
1
Allele Number *:
251462
Allele Frequency *:
0.000004
Variant Comments & Reference:
This alteration is predicted to affect pre-mRNA splicing and is a synonymous codon change (GGG_GGT) at codon G342 in exon 9. Variant creates a new donor splice site and introduces a premature stop codon at codon 348. Variant is believed to be a polymorphism. Ventura et al 2000Structural Interpretation:
Structural analysis cannot be performed on this (Point | Silent) variant. c.1033A>T
p.Lys345* (Legacy AA No. 327)
Variant Type:
Point
Domain:
Apple 4
Codon Change:
A>T
Variant Effect:
Nonsense
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Lin et al 2020Structural Interpretation:
Structural analysis cannot be performed on this (Point | Nonsense) variant. c.1060G>A
p.Gly354Arg (Legacy AA No. 336)
Variant Type:
Point
Domain:
Apple 4
Codon Change:
G>A
Variant Effect:
Missense
No. of Patients Reported:
2
Phenotype:
I
Allele Count *:
3
Allele Number *:
251410
Allele Frequency *:
0.000012
Variant Comments & Reference:
Saunders et al 2005Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1077A>G
p.Ile359Met (Legacy AA No. 341)
Variant Type:
Point
Domain:
Apple 4
Codon Change:
A>G
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
1
Allele Number *:
251412
Allele Frequency *:
0.000004
Variant Comments & Reference:
Ile359 is one of several residues involved in the non-covalent interactions between two Ap4 domains in a FXI dimer. Saunders et al 2009Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1079T>C
p.Leu360Pro (Legacy AA No. 342)
Variant Type:
Point
Domain:
Apple 4
Codon Change:
T>C
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
This non-conservative change in the surface region of the Apple 4 domain, where active FXII attaches, converts hydrophobic Leu to Pro. Substitution may lead to changes in the contact surface conformation, preventing appropriate activation of FXI. Fard-Esfahani et al 2008Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1084G>A
p.Gly362Arg (Legacy AA No. 344)
Variant Type:
Point
Domain:
Apple 4
Codon Change:
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
22
Allele Number *:
282750
Allele Frequency *:
0.000078
Variant Comments & Reference:
Saunders et al 2009Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1102G>A
p.Gly368Arg (Legacy AA No. 350)
Variant Type:
Point
Domain:
Apple 4
Codon Change:
G>A
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
I
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
The Gly368Arg mutant is thought not to be secreted as the patient in which the mutation was found shows a parallel defect of both FXI:C and FXI:Ag levels. Quelin et al 2009Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1103G>C
p.Gly368Ala (Legacy AA No. 350)
Variant Type:
Point
Domain:
Apple 4
Codon Change:
G>C
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
II
Allele Count *:
1
Allele Number *:
251366
Allele Frequency *:
0.000004
Variant Comments & Reference:
Decreases FXI dimerisation. Saunders et al 2009Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1103G>A
p.Gly368Glu (Legacy AA No. 350)
Variant Type:
Point
Domain:
Apple 4
Codon Change:
G>A
Variant Effect:
Missense
No. of Patients Reported:
4
Phenotype:
I
Allele Count *:
3
Allele Number *:
251366
Allele Frequency *:
0.000012
Variant Comments & Reference:
FXI-Gly368Glu (FXI-Nagoya II), with a more profound defect in dimerisation than FXI-Phe301Leu, would not be expected to influence secretion of wild-type protein appreciably, as was observed in co-transfection experiments. Northern blot analysis demonstrated that wild type and mutant protein generated similar amounts of mRNA. Kravtsov et al 2004, Yang et al 2021Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1106A>C
p.Tyr369Ser (Legacy AA No. 351)
Variant Type:
Point
Domain:
Apple 4
Codon Change:
A>C
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Tyr369 is highly conserved in the FXI amino acid sequences of related proteases of different species, indicating the importance of this residue to the structure of Ap4 domain in FXIa. Ap4 is important for the dimerisation of FXIa. A missense substitution at the adjoining residue Gly368Glu (factor XI Nagoya II) has been demonstrated (by expression studies) to have a profound effect on FXIa dimerization. It is possible that Tyr369Ser could have a similar effect on FXIa dimerisation. Kravtsov et al 2004, Jayandharan et al 2005Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1107C>A
p.Tyr369* (Legacy AA No. 351)
Variant Type:
Point
Domain:
Apple 4
Codon Change:
C>A
Variant Effect:
Nonsense
No. of Patients Reported:
9
Phenotype:
U
Allele Count *:
10
Allele Number *:
282746
Allele Frequency *:
0.000035
Variant Comments & Reference:
Saunders et al 2005, Zhang et al 2020Structural Interpretation:
Structural analysis cannot be performed on this (Point | Nonsense) variant. c.1107C>T
p.Tyr369= (Legacy AA No. 351)
Variant Type:
Point
Domain:
Apple 4
Codon Change:
C>T
Variant Effect:
Silent
No. of Patients Reported:
3
Phenotype:
U
Allele Count *:
1
Allele Number *:
251352
Allele Frequency *:
0.000004
Variant Comments & Reference:
Lin et al 2020, Shao et al 2016Structural Interpretation:
Structural analysis cannot be performed on this (Point | Silent) variant. c.1118T>C
p.Leu373Ser (Legacy AA No. 355)
Variant Type:
Point
Domain:
Apple 4
Codon Change:
T>C
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
9
Allele Number *:
282678
Allele Frequency *:
0.000032
Variant Comments & Reference:
May disrupt a neighbouring disulphide bridge. Saunders et al 2009Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1120T>C
p.Cys374Arg (Legacy AA No. 356)
Variant Type:
Point
Domain:
Apple 4
Codon Change:
T>C
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Disrupts Cys291-Cys374 disulphide bridge. Saunders et al 2009Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1135+5G>A
(Legacy AA No. 360)
Variant Type:
Point
Domain:
Intronic
Codon Change:
G>A
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
6
Allele Number *:
281714
Allele Frequency *:
0.000021
Variant Comments & Reference:
Saunders et al 2009Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1165G>A
p.Val389Ile (Legacy AA No. 371)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
G>A
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
II
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Residue Val389 is located one residue following the cleavage site of FXI (P2' position, according to the convention of numbering position around the scissile bond), thus is part of the activation loop. FXI antigen levels, measured in both conditioned media and lysates of cells expressing the mutant protein (in either the heterozygous or homozygous state), were not significantly different from those measured in wild-type samples. FXI specific activity was measured in conditioned media. The specific activity of the FXI Val389Ile protein was significantly reduced when compared with the wildtype one (30% and 80% for the heterozygous and the homozygous conditions, respectively). Given the proximity of Val389 to the FXI activation site, a possible interference on FXI activation was hypothesized and this mutation was found to be associated with a defect both in FXI activation (slower than normal), and in FIX activation (slightly delayed), thus supporting the role of residues neighboring the active site in influencing and stabilizing the enzyme active state. Bozzao et al 2007Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1169G>C
p.Gly390Ala (Legacy AA No. 372)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
G>C
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Karimi et al 2009Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1178C>T
p.Ala393Val (Legacy AA No. 375)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
C>T
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
II
Allele Count *:
2
Allele Number *:
251490
Allele Frequency *:
0.000008
Variant Comments & Reference:
Ala393Val is exposed in the SP domain on the opposite side of where the His431-Asp480-Ser575 catalytic triad at the active site is located; however, its effect is not clear from the FXI structure. Saunders et al 2009Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1186C>T
p.Arg396Cys (Legacy AA No. 378)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
C>T
Variant Effect:
Missense
No. of Patients Reported:
2
Phenotype:
II
Allele Count *:
5
Allele Number *:
251486
Allele Frequency *:
0.000020
Variant Comments & Reference:
Expression studies indicate that this is a causative mutation (rather than a benign polymorphism -Germanos-Haddad et al 2003). FXI antigen assays confirm that FXI-Cys396 is secreted. FXI activity assays show FXI-Cys396 to have minimal activity, demonstrating that it is functionally inactive in an APTT-based assay. If, as is suggested in the Germanos-Haddad abstract, Arg396Cys is identified in the 'normal' lebanese population, thus leading to it being misassigned as a polymorphism, then it is possible that it is a common founder mutation. Mitchell et al 2007Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1191T>C
p.Gly397= (Legacy AA No. 379)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
T>C
Variant Effect:
Silent
No. of Patients Reported:
0
Phenotype:
None
Allele Count *:
42597
Allele Number *:
282848
Allele Frequency *:
0.150600
Variant Comments & Reference:
Frequency in a random sample of normal volunteers is 18%, making this variant a polymorphism. Martincic et al 1998, Wiewel-Verschueren et al 2017Structural Interpretation:
Structural analysis cannot be performed on this (Point | Silent) variant. c.1195T>C
p.Trp399Arg (Legacy AA No. 381)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
T>C
Variant Effect:
Missense
No. of Patients Reported:
0
Phenotype:
None
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Polymorphism. Unpublished DataStructural Interpretation:
Please click HERE for in-depth variant analysis. C.1196G>T
p.Trp399Leu (Legacy AA No. 381)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
G>T
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
I
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Variant likely leads to altered protein secretion. It is unlikely to have an effect on the structure of the catalytic domain as both Trp and Leu are hydrophobic amino acids. Quelin et al 2009Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1199C>T
p.Pro400Leu (Legacy AA No. 382)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
C>T
Variant Effect:
Missense
No. of Patients Reported:
3
Phenotype:
I
Allele Count *:
2
Allele Number *:
251476
Allele Frequency *:
0.000008