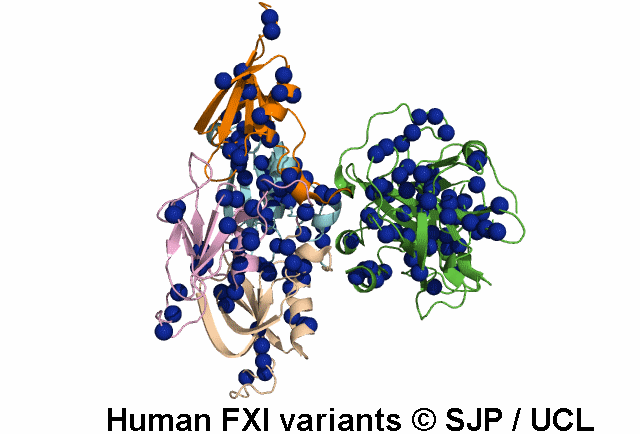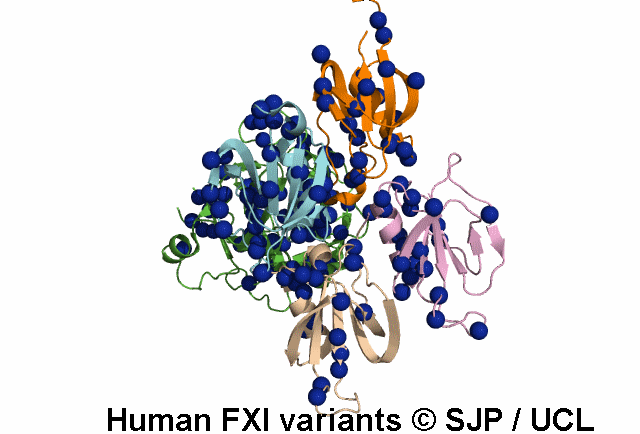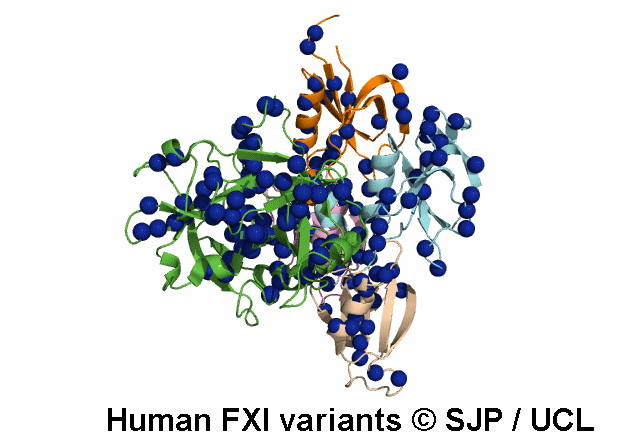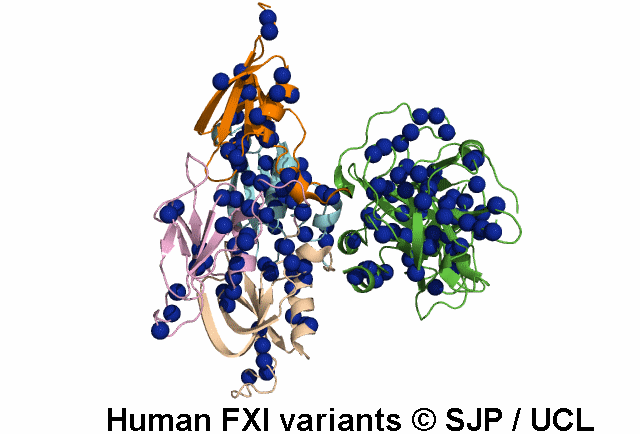
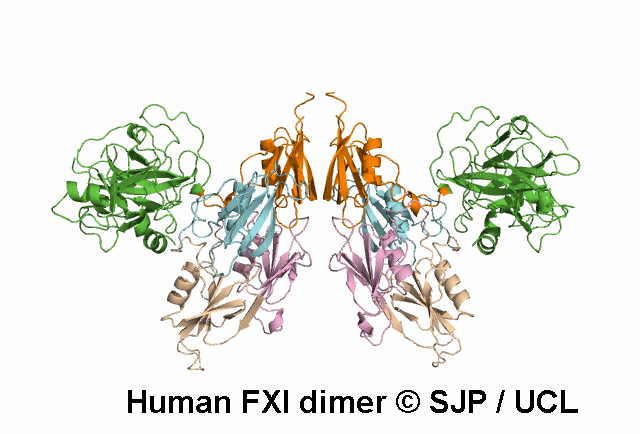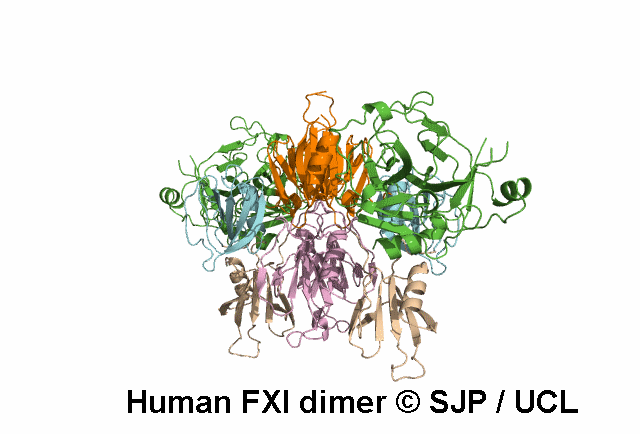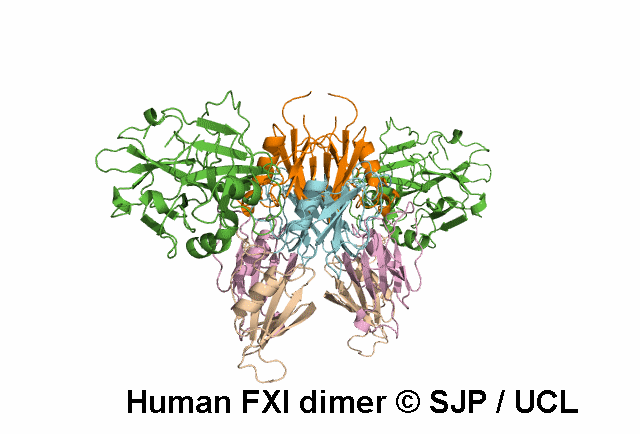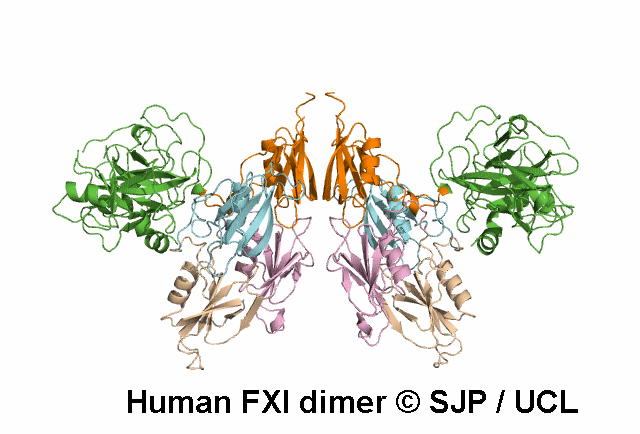Data Export Options:
Full List of Variants: 272 unique variants retrieved (displaying 50 entries per page). Scroll down to navigate to the next page(s).
Terms with a '*' next to them are explained on the Help Page .
c.1028+5G>T
-
Variant Type:
Point
Domain:
Apple 4
Codon Change:
G>T
Variant Effect:
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
1
Allele Number *:
251452
Allele Frequency *:
0.000004
Variant Comments & Reference:
This FXI mutation abolishes the splice site at the 3' boundary of exon 9. Saunders et al 2009Structural Interpretation:
Structural analysis cannot be performed on this (Point | ) variant. c.1029-2A>G
-
Variant Type:
Point
Domain:
Apple 4
Codon Change:
A>G
Variant Effect:
No. of Patients Reported:
0
Phenotype:
I
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Pugh et al 1995Structural Interpretation:
Structural analysis cannot be performed on this (Point | ) variant. c.1072delA
-
Variant Type:
Deletion
Domain:
Apple 4
Codon Change:
delA
Variant Effect:
Frameshift
No. of Patients Reported:
2
Phenotype:
I
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Saunders et al 2005Structural Interpretation:
Structural analysis cannot be performed on this (Deletion | Frameshift) variant. c.1135+1G>A
-
Variant Type:
Point
Domain:
Intronic
Codon Change:
G>A
Variant Effect:
No. of Patients Reported:
2
Phenotype:
U
Allele Count *:
3
Allele Number *:
250710
Allele Frequency *:
0.000012
Variant Comments & Reference:
Mutation changes an invariant nucleotide of the splicing site recognition sequence, and likely results in aberrant splicing of FXI mRNA causing FXI deficiency. Okumura et al 2006Structural Interpretation:
Structural analysis cannot be performed on this (Point | ) variant. c.1136-4delGTTG
-
Variant Type:
Deletion
Domain:
Intronic
Codon Change:
delGTTG
Variant Effect:
Frameshift
No. of Patients Reported:
8
Phenotype:
I
Allele Count *:
1
Allele Number *:
251472
Allele Frequency *:
0.000004
Variant Comments & Reference:
Xie et al 2005, Zhang et al 2020Structural Interpretation:
Structural analysis cannot be performed on this (Deletion | Frameshift) variant. c.1304+12G>A
-
Variant Type:
Point
Domain:
Intronic
Codon Change:
G>A
Variant Effect:
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
876
Allele Number *:
277074
Allele Frequency *:
0.003162
Variant Comments & Reference:
Intron 11. Saunders et al 2009Structural Interpretation:
Structural analysis cannot be performed on this (Point | ) variant. c.1305-10T>A
-
Variant Type:
Point
Domain:
Intronic
Codon Change:
T>A
Variant Effect:
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
This mutation, in the polypyrimidine track of the intron K acceptor splice site affects FXI pre-mRNA splicing. It abolishes an AluI restriction site. This creates an alternative splice sites that results in the inclusion of intronic, or deletion of exonic nucleotides leading to frameshift. Ventura et al 2000Structural Interpretation:
Structural analysis cannot be performed on this (Point | ) variant. c.1322delT
-
Variant Type:
Deletion
Domain:
Serine Protease
Codon Change:
delT
Variant Effect:
Frameshift
No. of Patients Reported:
2
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Lin et al 2020Structural Interpretation:
Structural analysis cannot be performed on this (Deletion | Frameshift) variant. c.1325delT
p.Leu442Cys-
Variant Type:
Deletion
Domain:
Serine Protease
Codon Change:
delT
Variant Effect:
Frameshift
No. of Patients Reported:
0
Phenotype:
U
Allele Count *:
14
Allele Number *:
282776
Allele Frequency *:
0.000050
Variant Comments & Reference:
Induces formation of premature termination codon. Shao et al 2016Structural Interpretation:
Structural analysis cannot be performed on this (Deletion | Frameshift) variant. c.1448delT
-
Variant Type:
Deletion
Domain:
Serine Protease
Codon Change:
delT
Variant Effect:
Frameshift
No. of Patients Reported:
0
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Induces formation of premature termination codon. Shao et al 2016Structural Interpretation:
Structural analysis cannot be performed on this (Deletion | Frameshift) variant. c.1480+3A>T
-
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
A>T
Variant Effect:
Missense
No. of Patients Reported:
0
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Shao et al 2016Structural Interpretation:
Structural analysis cannot be performed on this (Point | Missense) variant. c.1481-215C>T
-
Variant Type:
Point
Domain:
Intronic
Codon Change:
C>T
Variant Effect:
No. of Patients Reported:
0
Phenotype:
None
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Polymorphism. Wiewel-Verschueren et al 2017Structural Interpretation:
Structural analysis cannot be performed on this (Point | ) variant. c.1481-34G>T
-
Variant Type:
Point
Domain:
Intronic
Codon Change:
G>T
Variant Effect:
No. of Patients Reported:
0
Phenotype:
None
Allele Count *:
17359
Allele Number *:
282360
Allele Frequency *:
0.061478
Variant Comments & Reference:
Polymorphism. Wiewel-Verschueren et al 2017Structural Interpretation:
Structural analysis cannot be performed on this (Point | ) variant. c.1481-188C>T
-
Variant Type:
Point
Domain:
Intronic
Codon Change:
C>T
Variant Effect:
No. of Patients Reported:
0
Phenotype:
None
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Polymorphism. Wiewel-Verschueren et al 2017Structural Interpretation:
Structural analysis cannot be performed on this (Point | ) variant. c.1556insG
-
Variant Type:
Insertion
Domain:
Serine Protease
Codon Change:
insG
Variant Effect:
Frameshift
No. of Patients Reported:
7
Phenotype:
I
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Saunders et al 2005Structural Interpretation:
Structural analysis cannot be performed on this (Insertion | Frameshift) variant. c.1560dupG
-
Variant Type:
Duplication
Domain:
Serine Protease
Codon Change:
dupG
Variant Effect:
Frameshift
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Mutation inserts an additional G in a stretch of five guanine nucleotides at nucleotide position 1560. This leads to a frameshift, generating a premature termination codon at position 535 [Tyr503ValfsX32]. Kwon et al 2008Structural Interpretation:
Structural analysis cannot be performed on this (Duplication | Frameshift) variant. c.1574-93dupAT
-
Variant Type:
Duplication
Domain:
Intronic
Codon Change:
dupAT
Variant Effect:
Frameshift
No. of Patients Reported:
0
Phenotype:
None
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Polymorphism. Peretz et al 1997Structural Interpretation:
Structural analysis cannot be performed on this (Duplication | Frameshift) variant. c.1575_1576delAG
-
Variant Type:
Deletion
Domain:
Serine Protease
Codon Change:
delAG
Variant Effect:
Frameshift
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Zhang et al 2020Structural Interpretation:
Structural analysis cannot be performed on this (Deletion | Frameshift) variant. c.1576+51C>A
-
Variant Type:
Point
Domain:
Intronic
Codon Change:
G>T
Variant Effect:
No. of Patients Reported:
0
Phenotype:
None
Allele Count *:
17287
Allele Number *:
281500
Allele Frequency *:
0.061410
Variant Comments & Reference:
Polymorphism. Wiewel-Verschueren et al 2017Structural Interpretation:
Structural analysis cannot be performed on this (Point | ) variant. c.1714_1716+11del
-
Variant Type:
Deletion
Domain:
Serine Protease
Codon Change:
Variant Effect:
No. of Patients Reported:
1
Phenotype:
I
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Jewish Type IV. Peretz et al 1996Structural Interpretation:
Structural analysis cannot be performed on this (Deletion | ) variant. c.1716+1G>A
-
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
G>A
Variant Effect:
No. of Patients Reported:
2
Phenotype:
I
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Jewish Type I mutation. Saunders et al 2005, Peretz et al 2013Structural Interpretation:
Structural analysis cannot be performed on this (Point | ) variant. c.1716+248G>A
-
Variant Type:
Point
Domain:
Intronic
Codon Change:
G>A
Variant Effect:
No. of Patients Reported:
2
Phenotype:
None
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Polymorphism. Wiewel-Verschueren et al 2017Structural Interpretation:
Structural analysis cannot be performed on this (Point | ) variant. c.1717-48A>G
-
Variant Type:
Point
Domain:
Intronic
Codon Change:
A>G
Variant Effect:
No. of Patients Reported:
0
Phenotype:
None
Allele Count *:
17467
Allele Number *:
281898
Allele Frequency *:
0.061962
Variant Comments & Reference:
Polymorphism. Cargill et al 1999Structural Interpretation:
Structural analysis cannot be performed on this (Point | ) variant. c.1717-1G>A
-
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
G>A
Variant Effect:
Missense
No. of Patients Reported:
0
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Zhang et al 2020Structural Interpretation:
Structural analysis cannot be performed on this (Point | Missense) variant. c.3G>T
p.Met1Ile (Legacy AA No. -18)
Variant Type:
Point
Domain:
Signal Peptide
Codon Change:
G>T
Variant Effect:
Missense
No. of Patients Reported:
3
Phenotype:
I
Allele Count *:
1
Allele Number *:
31402
Allele Frequency *:
0.000032
Variant Comments & Reference:
Mutations affecting the ATG initiation codon have not been widely reported, and the loss of the initiation methionine in the Met1Ile variant was expected to be severely detrimental to protein synthesis and secretion. Expression studies show that no FXI is secreted, but surprisingly apparently normal (160 kDa) dimeric FXI was detected by western blot from cell lysates. Mitchell et al 2007Structural Interpretation:
Please click HERE for in-depth variant analysis. c.15T>A
p.Tyr5* (Legacy AA No. -14)
Variant Type:
Point
Domain:
Signal Peptide
Codon Change:
T>A
Variant Effect:
Nonsense
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Neerman-Arbez et al 2007Structural Interpretation:
Structural analysis cannot be performed on this (Point | Nonsense) variant. c.44C>T
p.Ser15Leu (Legacy AA No. -4)
Variant Type:
Point
Domain:
Signal Peptide
Codon Change:
C>T
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
I
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Quelin et al 2006Structural Interpretation:
Please click HERE for in-depth variant analysis. c.52G>C
p.Gly18Arg (Legacy AA No. -1)
Variant Type:
Point
Domain:
Signal Peptide
Codon Change:
G>C
Variant Effect:
Missense
No. of Patients Reported:
0
Phenotype:
I
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Signal peptide prediction analysis results showed that this mutation would disrupt the original signal peptide cutting site between -1 and +1 amino acid position, resulting in impaired secretion of the synthesised FXI protein. Wang et al 2009Structural Interpretation:
Please click HERE for in-depth variant analysis. c.55G>T
p.Glu19* (Legacy AA No. 1)
Variant Type:
Point
Domain:
Linker
Codon Change:
G>T
Variant Effect:
Nonsense
No. of Patients Reported:
1
Phenotype:
I
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Mitchell et al 2006Structural Interpretation:
Structural analysis cannot be performed on this (Point | Nonsense) variant. c.55G>A
(Legacy AA No. 1)
Variant Type:
Point
Domain:
Linker
Codon Change:
G>A
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Zhang et al 2020Structural Interpretation:
Please click HERE for in-depth variant analysis. c.67C>T
p.Gln23* (Legacy AA No. 5)
Variant Type:
Point
Domain:
Apple 1
Codon Change:
C>T
Variant Effect:
Nonsense
No. of Patients Reported:
0
Phenotype:
U
Allele Count *:
3
Allele Number *:
251420
Allele Frequency *:
0.000012
Variant Comments & Reference:
Castaman et al 2007 (ISTH Abstract)Structural Interpretation:
Structural analysis cannot be performed on this (Point | Nonsense) variant. c.100G>C
p.Asp34His (Legacy AA No. 16)
Variant Type:
Point
Domain:
Apple 1
Codon Change:
G>C
Variant Effect:
Missense
No. of Patients Reported:
0
Phenotype:
I
Allele Count *:
2
Allele Number *:
251446
Allele Frequency *:
0.000008
Variant Comments & Reference:
Amount of mutant protein secreted from cells in vitro is reduced. The replacement of Asp 34 in the Ap1 domain of factor XI is a radical substitution, because the wild-type Asp side chain is small and negatively charged at physiologic pH, whereas the His group is bulky and hydrophobic with neutral charge - the alteration of charge and shape and the resultant conformational change of this region of the molecule might interfere with chain folding. Pugh et al 1995Structural Interpretation:
Please click HERE for in-depth variant analysis. c.113T>C
p.Val38Ala (Legacy AA No. 20)
Variant Type:
Point
Domain:
Apple 1
Codon Change:
T>C
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
I
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Substitution of FXI-Val38 by Ala in BHK cells caused profoundly decreased FXI secretion (22% of wild-type by cells) despite the presence of only slightly reduced amounts of FXI dimer within the cells. Zivelin et al 1999 (Abstract 2), Zucker et al 2007Structural Interpretation:
Please click HERE for in-depth variant analysis. c.122C>T
p.Pro41Leu (Legacy AA No. 23)
Variant Type:
Point
Domain:
Apple 1
Codon Change:
C>T
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
I
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Quelin et al 2005Structural Interpretation:
Please click HERE for in-depth variant analysis. c.122C>A
p.Pro41Gln (Legacy AA No. 23)
Variant Type:
Point
Domain:
Apple 1
Codon Change:
C>A
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Mitchell et al 2006Structural Interpretation:
Please click HERE for in-depth variant analysis. c.126C>G
p.Ser42Arg (Legacy AA No. 24)
Variant Type:
Point
Domain:
Apple 1
Codon Change:
C>G
Variant Effect:
Missense
No. of Patients Reported:
2
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Ser42Arg lies at the start of helix A1 within the Ap1 domain. In the crystal structure, this sidechain points towards the interdomain contacts with the Ap2 domain and the replacement with a larger charged residue is predicted to affect the protein structure. Saunders et al 2009Structural Interpretation:
Please click HERE for in-depth variant analysis. c.127G>A
p.Ala43Thr (Legacy AA No. 25)
Variant Type:
Point
Domain:
Apple 1
Codon Change:
Variant Effect:
Missense
No. of Patients Reported:
0
Phenotype:
U
Allele Count *:
9
Allele Number *:
251460
Allele Frequency *:
0.000036
Variant Comments & Reference:
Heterodimer trapping. Duga & Salomon 2013Structural Interpretation:
Please click HERE for in-depth variant analysis. c.137G>T
p.Cys46Phe (Legacy AA No. 28)
Variant Type:
Point
Domain:
Apple 1
Codon Change:
G>T
Variant Effect:
Missense
No. of Patients Reported:
4
Phenotype:
I
Allele Count *:
3
Allele Number *:
251460
Allele Frequency *:
0.000012
Variant Comments & Reference:
This mutation represents a structurally significant change as it abolishes a Cys46-Cys76 disulphide bridge that is essential for the folding of the first apple domain. Zivelin et al 2002, Hill et al 2005Structural Interpretation:
Please click HERE for in-depth variant analysis. c.141G>C
p.Gln47His (Legacy AA No. 29)
Variant Type:
Point
Domain:
Apple 1
Codon Change:
G>C
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
I
Allele Count *:
17
Allele Number *:
251470
Allele Frequency *:
0.000068
Variant Comments & Reference:
Mutant protein was not secreted by transfected HEK293 cells. Bolton-Maggs et al 2003 (Abstract)Structural Interpretation:
Please click HERE for in-depth variant analysis. c.151A>C
p.Thr51Pro (Legacy AA No. 33)
Variant Type:
Point
Domain:
Apple 1
Codon Change:
A>C
Variant Effect:
Missense
No. of Patients Reported:
2
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Thr51Pro and Thr51Ile. These changes convert hydrophilic and moderately sized threonine to hydrophobic, large proline and isoleucine, respectively. Thr51 lies in the first a-helix, in a buried region of Apple 1 domain and just next to a cysteine residue which forms the disulphide bond. On the other hand, the threonine residue is conserved in positions 51, 141, 231 and 322 of four Apple domains, in the same position relative to the a-helix of each domain. Therefore, it seems that this amino acid is essential for proper function of the protein and its alteration is not conservative. Fard-Esfahani et al 2008, Colakoglu et al 2018Structural Interpretation:
Please click HERE for in-depth variant analysis. c.152C>T
p.Thr51Ile (Legacy AA No. 33)
Variant Type:
Point
Domain:
Apple 1
Codon Change:
C>T
Variant Effect:
Missense
No. of Patients Reported:
0
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Thr51Pro and Thr51Ile. These changes convert hydrophilic and moderately sized threonine to hydrophobic, large proline and isoleucine, respectively. Thr51 lies in the first a-helix, in a buried region of Apple 1 domain and just next to a cysteine residue which forms the disulphide bond. On the other hand, the threonine residue is conserved in positions 51, 141, 231 and 322 of four Apple domains, in the same position relative to the a-helix of each domain. Therefore, it seems that this amino acid is essential for proper function of the protein and its alteration is not conservative. Fard-Esfahani et al 2008Structural Interpretation:
Please click HERE for in-depth variant analysis. c.166T>C
p.Cys56Arg (Legacy AA No. 38)
Variant Type:
Point
Domain:
Apple 1
Codon Change:
T>C
Variant Effect:
Missense
No. of Patients Reported:
36
Phenotype:
I
Allele Count *:
3
Allele Number *:
251464
Allele Frequency *:
0.000012
Variant Comments & Reference:
High prevalence in French Basque population. Mutant protein synthesized in BHK transfected cells but not secreted. Zivelin et al 2002, Esteban et al 2017Structural Interpretation:
Please click HERE for in-depth variant analysis. c.168T>A
p.Cys56* (Legacy AA No. 38)
Variant Type:
Point
Domain:
Apple 1
Codon Change:
T>A
Variant Effect:
Nonsense
No. of Patients Reported:
0
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Ramadan et al 2006Structural Interpretation:
Structural analysis cannot be performed on this (Point | Nonsense) variant. c.168T>G
p.Cys56Trp (Legacy AA No. 38)
Variant Type:
Point
Domain:
Apple 1
Codon Change:
T>G
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
I
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Cys56 is a buried residue located in a random coil of Ap1 and is involved in the Cys50-Cys56 disulphide bridge. This disulphide bridge represents one of three conserved bridges that are responsible for the correct folding of the Ap domains. Therefore, Cys56Trp mutation is likely to significantly impair folding of the Ap1 domain. Expression studies reveal a secretion defect. Castaman et al 2005, Spena et al 2009Structural Interpretation:
Please click HERE for in-depth variant analysis. c.197C>T
p.Pro66Leu (Legacy AA No. 48)
Variant Type:
Point
Domain:
Apple 1
Codon Change:
C>T
Variant Effect:
Missense
No. of Patients Reported:
0
Phenotype:
None
Allele Count *:
18
Allele Number *:
282848
Allele Frequency *:
0.000064
Variant Comments & Reference:
Polymorphism. Cargill et al 1999Structural Interpretation:
Please click HERE for in-depth variant analysis. c.209C>T
p.Pro70Leu (Legacy AA No. 52)
Variant Type:
Point
Domain:
Apple 1
Codon Change:
C>T
Variant Effect:
Missense
No. of Patients Reported:
0
Phenotype:
U
Allele Count *:
1
Allele Number *:
251440
Allele Frequency *:
0.000004
Variant Comments & Reference:
Unpublished Data From Coagulation factor XI: a database of mutations and polymorphism associated with factor XI deficiency http://www.wienkav.at/kav/factorxi/Faktor_XI_Suche.aspStructural Interpretation:
Please click HERE for in-depth variant analysis. c.214C>T
p.Arg72* (Legacy AA No. 54)
Variant Type:
Point
Domain:
Apple 1
Codon Change:
C>T
Variant Effect:
Nonsense
No. of Patients Reported:
0
Phenotype:
U
Allele Count *:
1
Allele Number *:
251432
Allele Frequency *:
0.000004
Variant Comments & Reference:
Castaman et al 2005Structural Interpretation:
Structural analysis cannot be performed on this (Point | Nonsense) variant. c.215G>C
p.Arg72Pro (Legacy AA No. 54)
Variant Type:
Point
Domain:
Apple 1
Codon Change:
G>C
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
I
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Arg72 is part of an elaborated turning loop connecting the fourth and fifth strands. Electrostatic interactions among Arg72, Glu68 and Asp69 help to maintain the loop architecture. The Arg72Pro mutation eliminates these interactions and results in three close Pro residues, making the loop quite rigid and altering its structure and electrostatic surface. Spena et al 2009Structural Interpretation:
Please click HERE for in-depth variant analysis. c.224C>T
p.Thr75Ile (Legacy AA No. 57)
Variant Type:
Point
Domain:
Apple 1
Codon Change:
C>T
Variant Effect:
Missense
No. of Patients Reported:
0
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Quelin et al 2009Structural Interpretation:
Please click HERE for in-depth variant analysis. c.226G>C
p.Cys76Arg (Legacy AA No. 58)
Variant Type:
Point
Domain:
Apple 1
Codon Change:
G>C
Variant Effect:
Missense
No. of Patients Reported:
0
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-