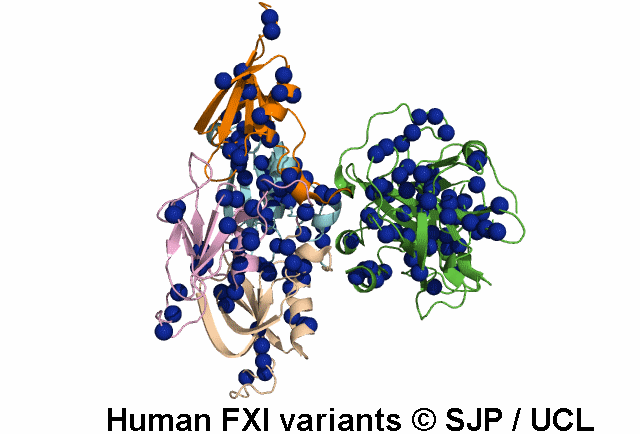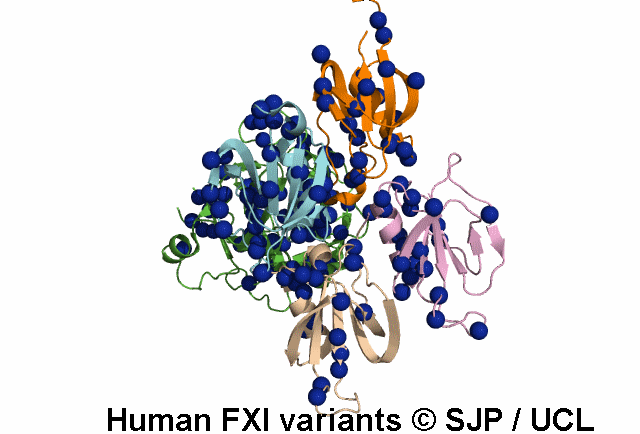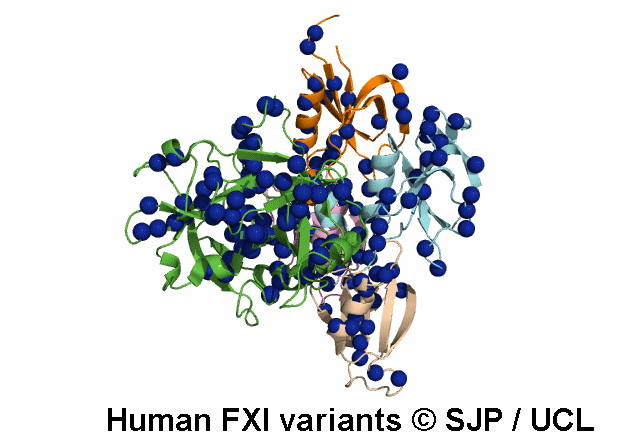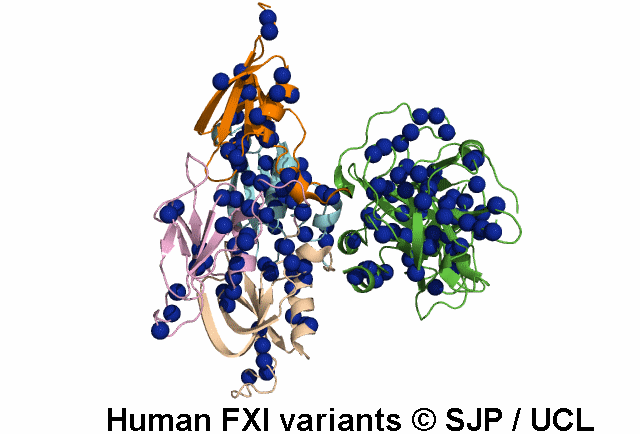
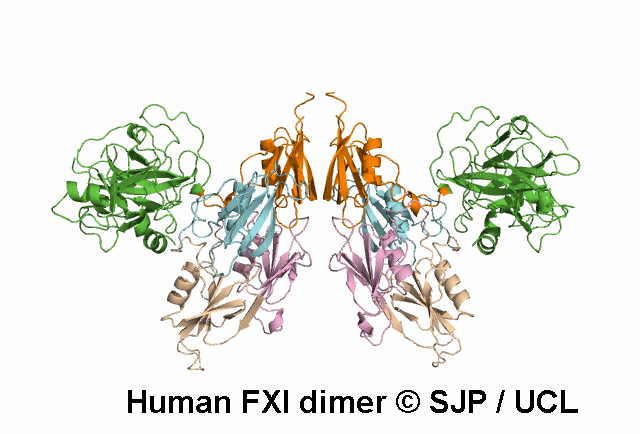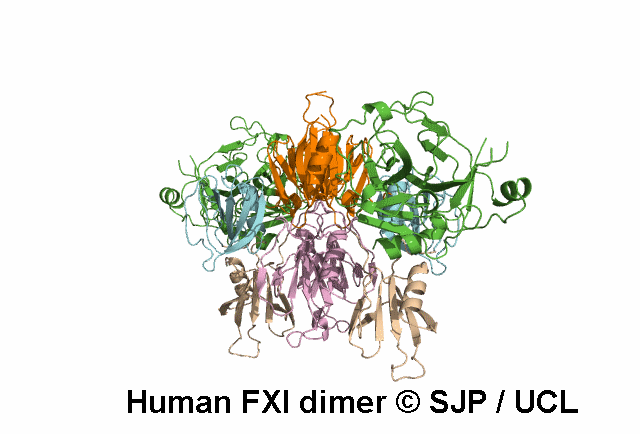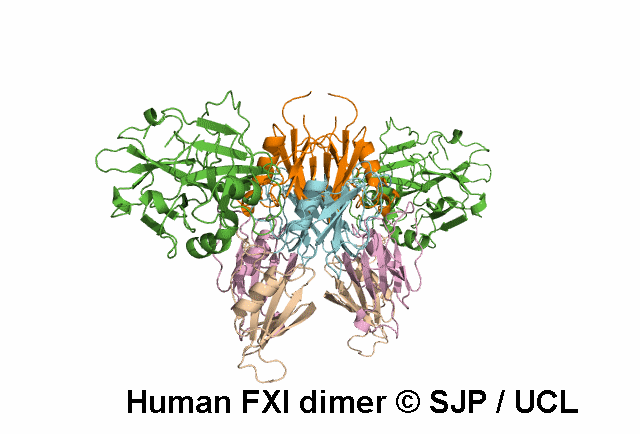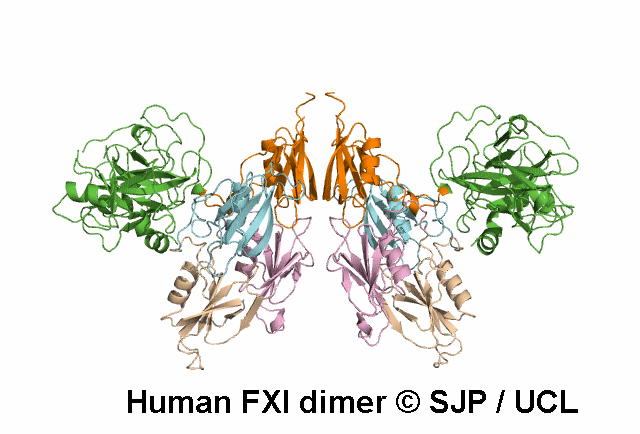Data Export Options:
Full List of Variants: 272 unique variants retrieved (displaying 50 entries per page). Scroll down to navigate to the next page(s).
Terms with a '*' next to them are explained on the Help Page .
c.1202G>A
p.Trp401* (Legacy AA No. 383)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
G>A
Variant Effect:
Nonsense
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Saunders et al 2009Structural Interpretation:
Structural analysis cannot be performed on this (Point | Nonsense) variant. Unspecified (V403M)
p.Val403Met (Legacy AA No. 385)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
Variant Effect:
Missense
No. of Patients Reported:
0
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Heterodimer trapping. Duga & Salomon 2013Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1211C>A
p.Thr404Asn (Legacy AA No. 386)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
C>A
Variant Effect:
Missense
No. of Patients Reported:
3
Phenotype:
I
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Saunders et al 2005Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1217A>C
p.His406Pro (Legacy AA No. 388)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
A>C
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
I
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
His406 is not charged but is involved in two polar contacts with Arg443 on the opposite beta strand. A turn induced by Pro is likely to disrupt the beta sheet structure. de Raucourt et al 2008Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1219A>C
p.Thr407Pro (Legacy AA No. 389)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
A>C
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Bolton-Maggs et al 2003 (Abstract)Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1222A>C
p.Thr408Pro (Legacy AA No. 390)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
A>C
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
2
Allele Number *:
251474
Allele Frequency *:
0.000008
Variant Comments & Reference:
Unpublished DataStructural Interpretation:
Please click HERE for in-depth variant analysis. c.1234C>T
p.Gln412* (Legacy AA No. 394)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
C>T
Variant Effect:
Nonsense
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Kawankar et al 2016Structural Interpretation:
Structural analysis cannot be performed on this (Point | Nonsense) variant. c.1247G>A
p.Cys416Tyr (Legacy AA No. 398)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
G>A
Variant Effect:
Missense
No. of Patients Reported:
17
Phenotype:
I
Allele Count *:
4
Allele Number *:
251468
Allele Frequency *:
0.000016
Variant Comments & Reference:
Mutant protein is not expressed by 293 human kidney fibroblasts. Western blot shows mutant protein forms intracellular dimer. Transfections show reduced secretion of mutant protein. Has a dominant negative effect on wild type secretion. This mutation breaks the disulphide bridge between Cys416-Cys432. Gailani et al 2001 (Abstract), Kravtsov et al 2005, Saunders et al 2009, Esteban et al 2017, Colakoglu et al 2018Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1252G>A
p.Gly418Ser (Legacy AA No. 400)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
G>A
Variant Effect:
Missense
No. of Patients Reported:
0
Phenotype:
U
Allele Count *:
1
Allele Number *:
251466
Allele Frequency *:
0.000004
Variant Comments & Reference:
Castaman et al 2007 (ISTH Abstract)Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1253G>T
p.Gly418Val (Legacy AA No. 400)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
G>T
Variant Effect:
Missense
No. of Patients Reported:
11
Phenotype:
I
Allele Count *:
2
Allele Number *:
251466
Allele Frequency *:
0.000008
Variant Comments & Reference:
Mutant protein is not expressed in 293 kidney fibroblasts. Northern blot analysis demonstrated that wild type and mutant protein generated similar amounts of mRNA. Co-transfection with wild-type and mutant protein reduces wild-type secretion about 50% - non-secretable mutant protein monomers trap wild-type polypeptides within cells through homodimer formation, resulting in lower FXI levels in plasma. Gly418Val exerts a dominant negative effect which stems from an impaired secretion of heterodimers consisting of a normal and a mutant monomer. Gailani 2001 et al (Abstract B), Kravtsov et al 2004, Colakoglu et al 2018, Peng et al 2020, Zhang et al 2020Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1255T>G
p.Ser419Ala (Legacy AA No. 401)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
T>G
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Hopmeier et al 2004Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1270C>T
p.Gln424* (Legacy AA No. 406)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
C>T
Variant Effect:
Nonsense
No. of Patients Reported:
2
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Saunders et al 2009, Shao et al 2016Structural Interpretation:
Structural analysis cannot be performed on this (Point | Nonsense) variant. Unspecified (W425L)
p.Trp425Leu (Legacy AA No. 407)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
Variant Effect:
Missense
No. of Patients Reported:
0
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Shao et al 2016Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1275G>C
p.Trp425Cys (Legacy AA No. 407)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
G>C
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
I
Allele Count *:
5
Allele Number *:
282812
Allele Frequency *:
0.000018
Variant Comments & Reference:
Kravtsov et al 2004, de Raucourt et al 2008Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1277T>C
p.Ile426Thr (Legacy AA No. 408)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
T>C
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Esteban et al 2017Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1283C>T
p.Thr428Ile (Legacy AA No. 410)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
C>T
Variant Effect:
Missense
No. of Patients Reported:
2
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Thr428 occupies a buried position within the FXI SP domain adjacent to His431 of the active site and may perturb either the folding or the function of the SP domain. Saunders et al 2009Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1288G>A
p.Ala430Thr (Legacy AA No. 412)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
G>A
Variant Effect:
Missense
No. of Patients Reported:
3
Phenotype:
I
Allele Count *:
20
Allele Number *:
282590
Allele Frequency *:
0.000071
Variant Comments & Reference:
Ala430 is located in an alpha helix portion. Substitution at this position with Thr is expected to disrupt the helix structure due to steric constraints from the neighbouring residues Phe433 and Tyr434. The introduction of a polar residue in place of the simple aliphatic side chain of Ala is likely to perturb the local folding of the protein. Homology modelling suggests that this substitution might impair FXI secretion by reducing the stability of the serine protease fold. de Raucourt et al 2008, Castaman et al 2008Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1288G>T
p.Ala430Ser (Legacy AA No. 412)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
G>T
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
I
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Ala430Ser occupies a buried position within the SP domain adjacent to His431 of the active site, and may perturb either the folding or function of the SP domain. Saunders et al 2009Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1289C>T
p.Ala430Val (Legacy AA No. 412)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
C>T
Variant Effect:
Missense
No. of Patients Reported:
2
Phenotype:
I
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Homology modelling suggests that this substitution may reduce the stability of the serine protease fold, resulting in non-secretion. O'Connell et al 2005Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1327C>T
p.Arg443Cys (Legacy AA No. 425)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
C>T
Variant Effect:
Missense
No. of Patients Reported:
2
Phenotype:
I
Allele Count *:
7
Allele Number *:
282750
Allele Frequency *:
0.000025
Variant Comments & Reference:
Saunders et al 2009Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1334A>G
p.Tyr445Cys (Legacy AA No. 427)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
A>G
Variant Effect:
Missense
No. of Patients Reported:
2
Phenotype:
I
Allele Count *:
1
Allele Number *:
251392
Allele Frequency *:
0.000004
Variant Comments & Reference:
Expressing Tyr445Cys in BHK cells resulted in abrogation of FXI secretion despite intact dimerisation, and was associated with a reduced amount of FXI in lysed cells. Zucker et al 2007Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1336A>G
p.Ser446Gly (Legacy AA No. 428)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
A>G
Variant Effect:
Missense
No. of Patients Reported:
0
Phenotype:
None
Allele Count *:
2
Allele Number *:
251392
Allele Frequency *:
0.000008
Variant Comments & Reference:
One small hydrophilic amino acid (Ser) is replaced by another small, hydrophilic amino acid (Gly). Ser446 is conserved in bovine FXI and human prekallikrein, but it is not conserved in rabbit or murine FXI. Likely to be a non-causative polymorphism. Duncan et al 2008Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1394C>G
p.Gln451Glu (Legacy AA No. 433)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
C>G
Variant Effect:
Missense
No. of Patients Reported:
4
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Expected to lead to disruption of the catalytic domain structure of FXI molecule. Gln451 is highly conserved among humans, cattle, mice and dogs, as well as red jungle fowl, which suggests that Gln451 may be crucial for FXI activity. Gln has no charge, while Glu has a net negative charge at physiological pH. The charge change during Gln to Glu substitution may disturb the electrostatic properties of FXI, leading to abnormal activity. Ishikawa et al 2007Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1694T>A
p.Ile454Lys (Legacy AA No. 436)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
T>A
Variant Effect:
Missense
No. of Patients Reported:
0
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Induces a large conformational change within the 450–460 loop. Girolami et al 2011Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1378T>G
p.Phe460Val (Legacy AA No. 442)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
T>G
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
I
Allele Count *:
1
Allele Number *:
251396
Allele Frequency *:
0.000004
Variant Comments & Reference:
Oligonucleotide probes of WT and mutant were hybridized to 50 normal individuals. Only WT hydrized - so not a polymorphism. Imanaka et al 1995Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1393G>T
p.Glu465* (Legacy AA No. 447)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
G>T
Variant Effect:
Nonsense
No. of Patients Reported:
1
Phenotype:
I
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
The G to T transversion in exon 12 results in a nonsense mutation Glu465* which leads to the disruption of the catalytic domain structure of the FXI molecule. Tsukahara et al 2003Structural Interpretation:
Structural analysis cannot be performed on this (Point | Nonsense) variant. c.1432G>A
p.Gly478Arg (Legacy AA No. 460)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
G>A
Variant Effect:
Missense
No. of Patients Reported:
11
Phenotype:
I
Allele Count *:
26
Allele Number *:
251454
Allele Frequency *:
0.000103
Variant Comments & Reference:
Mutant protein is expressed at levels comparable to normal FXI, and mutant protein was not secreted. Bolton-Maggs et al 2003 (Abstract)Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1448T>C
p.Leu483Ser (Legacy AA No. 465)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
T>C
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Kawankar et al 2016Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1478C>T
p.Thr493Ile (Legacy AA No. 475)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
C>T
Variant Effect:
Missense
No. of Patients Reported:
3
Phenotype:
I
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Expression of mutant protein shows mutant is secreted poorly compared to wild type. This mutation destroys an N-linked glycosylation site. Substitution of Thr493 with Ala, Pro, Lys or Arg all abolish the site and also severely reduce level of secreted FXI:Ag. Substitution with Ser which does not abolish the site had no affect on secretion. However substitution of Asn491 with Ala which abolishes the glycosylation site also had no affect on secretion. This indicates the cause of failure to secrete FXI is not the loss of glycolsylation site. McVey et al 2005Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1480+2T>G
(Legacy AA No. 476)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
T>G
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Zucker et al 2007Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1489C>T
p.Arg497* (Legacy AA No. 479)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
C>T
Variant Effect:
Nonsense
No. of Patients Reported:
2
Phenotype:
I
Allele Count *:
3
Allele Number *:
251402
Allele Frequency *:
0.000012
Variant Comments & Reference:
Saunders et al 2009Structural Interpretation:
Structural analysis cannot be performed on this (Point | Nonsense) variant. c.1498T>C
p.Cys500Arg (Legacy AA No. 482)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
T>C
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
2
Allele Number *:
251430
Allele Frequency *:
0.000008
Variant Comments & Reference:
Cys500Arg breaks the disulphide bridge between Cys500-Cys380. Saunders et al 2009Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1500C>G
p.Cys500Trp (Legacy AA No. 482)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
C>G
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
1
Allele Number *:
251420
Allele Frequency *:
0.000004
Variant Comments & Reference:
Cys500Trp breaks the disulphide bridge between Cys500-Cys380. Saunders et al 2009Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1507T>C
p.Ser503Pro (Legacy AA No. 485)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
T>C
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Ser503Pro converts hydrophilic small sized Ser to hydrophobic, large Pro in the buried region of the SP domain. Ser503 is in the random coil area of the structure, and may cause damaging structural effects. Fard-Esfahani et al 2008Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1531T>C
p.Tyr511His (Legacy AA No. 493)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
T>C
Variant Effect:
Missense
No. of Patients Reported:
2
Phenotype:
I
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Cells transfected with mutant protein contained reduced amounts of FXI and displayed decreased secretion. Zivelin et al 2002Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1545G>T
p.Trp515Cys (Legacy AA No. 497)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
G>T
Variant Effect:
Missense
No. of Patients Reported:
4
Phenotype:
I
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Saunders et al 2009, Tiscia et al 2017Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1546G>A
p.Val516Met (Legacy AA No. 498)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
G>A
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
7
Allele Number *:
251456
Allele Frequency *:
0.000028
Variant Comments & Reference:
Val516 is highly conserved across different species. The Val516Met mutation generates an ectopic Met residue close to the disulphide bond between C514 and C581. Bioinformatics studies using surface mapping of the evolutionary conservation level revealed that Val498 is a functionally important residue. Kwon et al 2008Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1556G>A
p.Trp519* (Legacy AA No. 501)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
G>A
Variant Effect:
Nonsense
No. of Patients Reported:
14
Phenotype:
I
Allele Count *:
8
Allele Number *:
251434
Allele Frequency *:
0.000032
Variant Comments & Reference:
Disrupts catalytic domain structure. Saunders et al 2005, Colakoglu et al 2018, Zhang et al 2020, Yang et al 2021Structural Interpretation:
Structural analysis cannot be performed on this (Point | Nonsense) variant. c.1556G>C
p.Trp519Ser (Legacy AA No. 501)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
G>C
Variant Effect:
Missense
No. of Patients Reported:
4
Phenotype:
I
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Variant likely deleterious. Shao et al 2016, Liu et al 2019Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1557G>C
p.Trp519Cys (Legacy AA No. 501)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
G>C
Variant Effect:
Missense
No. of Patients Reported:
4
Phenotype:
I
Allele Count *:
1
Allele Number *:
251432
Allele Frequency *:
0.000004
Variant Comments & Reference:
Saunders et al 2005Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1562A>G
p.Tyr521Cys (Legacy AA No. 503)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
A>G
Variant Effect:
Missense
No. of Patients Reported:
4
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Su et al 2018Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1608G>C
p.Lys536Asn (Legacy AA No. 518)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
G>C
Variant Effect:
Missense
No. of Patients Reported:
4
Phenotype:
I
Allele Count *:
1
Allele Number *:
251446
Allele Frequency *:
0.000004
Variant Comments & Reference:
The novel missense mutation FXI Lys536Asn leads to the replacement of the Lys536 which is part of a LxxxxxPxxxxxxC motif (x-variable amino acid) that occurs in several subfamilies of serine proteases. Although this motif is highly conserved between FXI and pre-kallikrein (86% sequence identity), the Lys536 is not conserved. On the other hand, this residue is conserved in human, murine and bovine FXI indicating that this is a FXI-specific amino acid. A molecular model of FXI predicts that the side-chain of this residue is on the surface of the molecule, possibly H-bonded to W515. Replacement of the positively charged lysine side chain by the smaller, neutral side chain of asparagine may interfere with the folding of the FXI molecule and/or secretion, leading to low antigen level. Alternatively, the secreted FXI variant may have a shortened plasma half-life. Ventura et al 2000, Esteban et al 2017Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1613C>T
p.Pro538Leu (Legacy AA No. 520)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
C>T
Variant Effect:
Missense
No. of Patients Reported:
9
Phenotype:
II
Allele Count *:
27
Allele Number *:
282666
Allele Frequency *:
0.000096
Variant Comments & Reference:
Mutant protein is expressed in 293 human kidney fibroblasts. Activated mutant has a modest catalytic defect in functional assays. The basis of catalytic defect that results from Pro538Leu alteration is due to subtle alterations in the oxyanion hole, which develops during conversion of zymogen to active enzyme, and results in a ~3.5-fold decrease in catalytic efficiency, consistent with the 70-80% loss of activity noted in conventional coagulation assays. Thus, Pro538 is important in maintaining the normal conformation of the FXI active site. Pro538 is conserved in FX and FVII, and its mutation in both these other proteins also causes Type II phenotypes.This variant was not activated by FXIIa. Gailani et al 2001 (Abstract), Gailani et al 2007, Saunders et al 2009, Esteban et al 2017Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1619T>G
p.Val540Gly (Legacy AA No. 522)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
T>G
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Affect the catalytic activity of activated FXI. Colakoglu et al 2018Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1634G>A
p.Cys545Tyr (Legacy AA No. 527)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
G>A
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
I
Allele Count *:
1
Allele Number *:
251462
Allele Frequency *:
0.000004
Variant Comments & Reference:
Expression of Cys545Tyr in BHK cells revealed intact dimerisation, a reduced amount of FXI in lysed cells and total lack of secretion from the cells. Zucker et al 2007Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1682G>A
p.Ala561Asp (Legacy AA No. 543)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
G>A
Variant Effect:
Missense
No. of Patients Reported:
2
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Damaging effect on FXI structure. Tiscia et al 2017Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1684G>A
p.Gly562Ser (Legacy AA No. 544)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
G>A
Variant Effect:
Missense
No. of Patients Reported:
2
Phenotype:
U
Allele Count *:
10
Allele Number *:
251490
Allele Frequency *:
0.000040
Variant Comments & Reference:
Gly562Ser resides on a surface exposed loop within the SP domain and it is not clear whether this is the causative mutation because the patients mother has the mutation but has normal FXI:C levels. Saunders et al 2009Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1693G>A
p.Glu565Lys (Legacy AA No. 547)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
G>A
Variant Effect:
Missense
No. of Patients Reported:
10
Phenotype:
I
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Responsible for an aberrant splicing in which exon 13 is skipped. Saunders et al 2009, Esteban et al 2017Structural Interpretation:
Please click HERE for in-depth variant analysis. c.1707C>T
p.Asp569= (Legacy AA No. 551)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
C>T
Variant Effect:
Silent
No. of Patients Reported:
0
Phenotype:
None
Allele Count *:
4190
Allele Number *:
282698
Allele Frequency *:
0.014821
Variant Comments & Reference:
Polymorphism. Cargill et al 1999Structural Interpretation:
Structural analysis cannot be performed on this (Point | Silent) variant. c.1717-2A>G
(Legacy AA No. 555)
Variant Type:
Point
Domain:
Serine Protease
Codon Change:
A>G
Variant Effect:
No. of Patients Reported:
0
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-