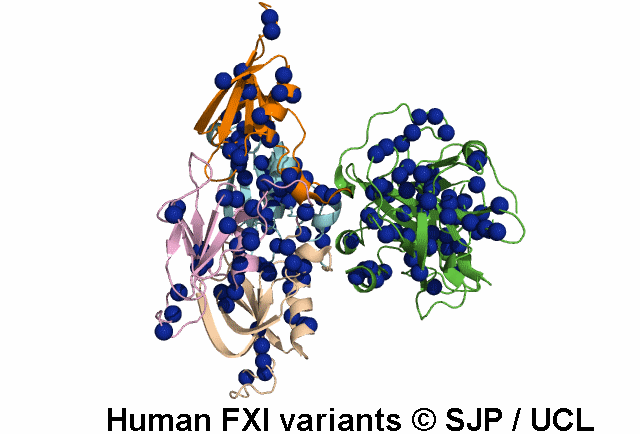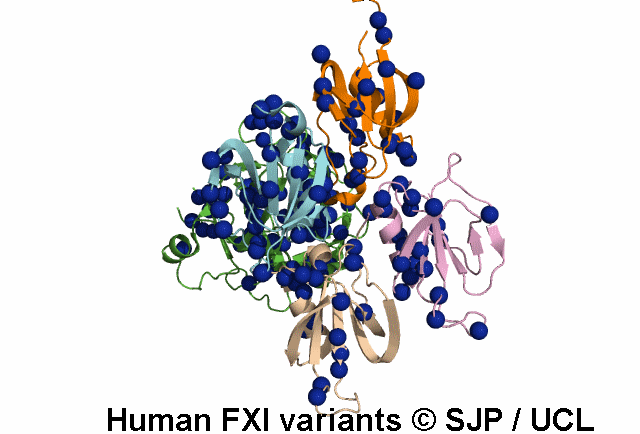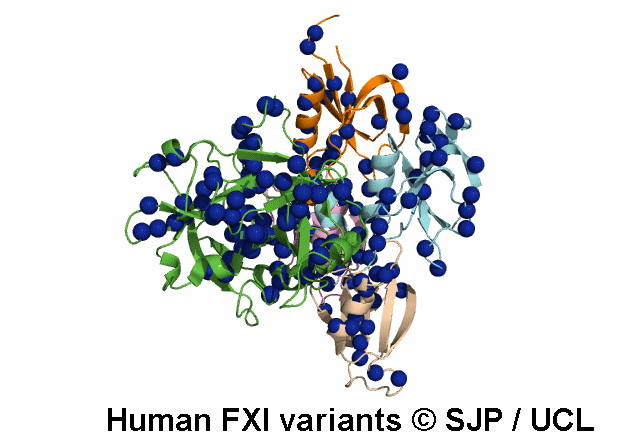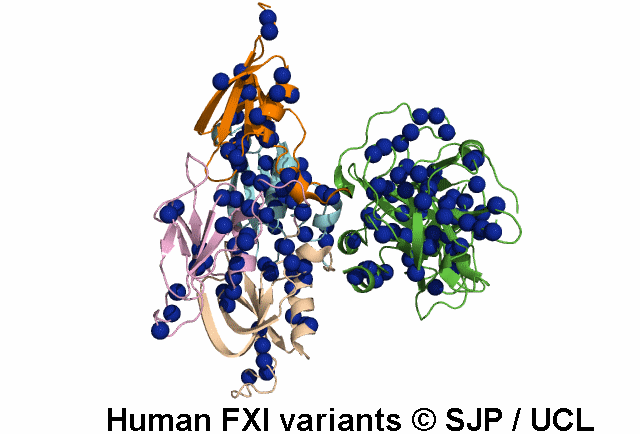
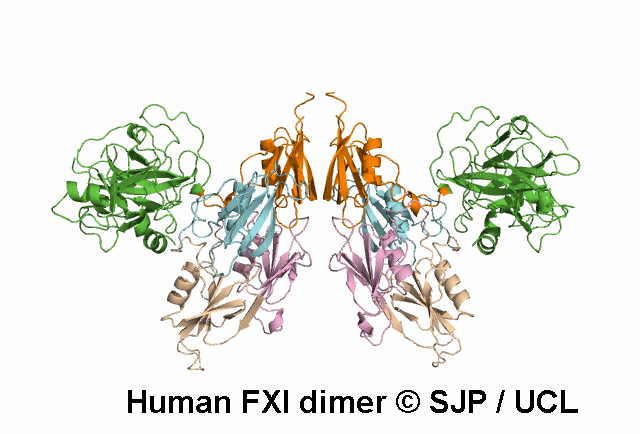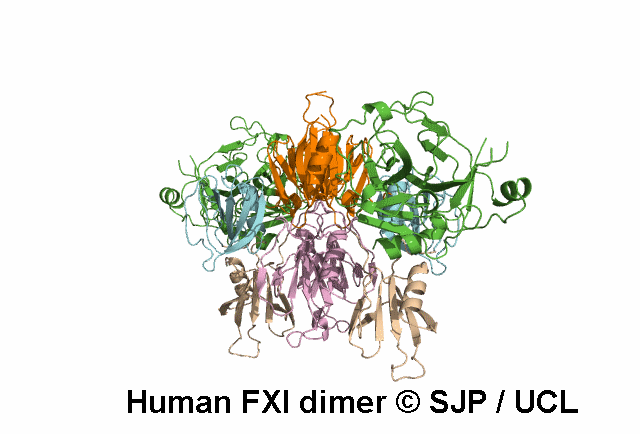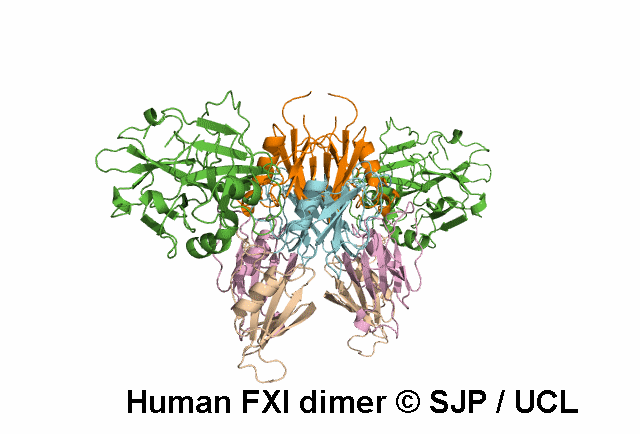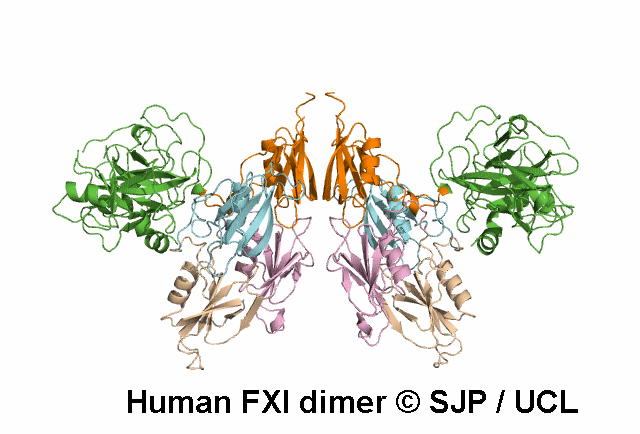Data Export Options:
Full List of Variants: 272 unique variants retrieved (displaying 50 entries per page). Scroll down to navigate to the next page(s).
Terms with a '*' next to them are explained on the Help Page .
c.227G>A
p.Cys76Tyr (Legacy AA No. 58)
Variant Type:
Point
Domain:
Apple 1
Codon Change:
G>A
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
I
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Abrogation of FXI secretion. This mutation disrupts the Cys76-Cys46 bond that is one of the three disulphide bonds responsible for correct folding of the Ap1 domain. Expression studies revealed intact FXI dimerization but no secretion at all from BHK cells. Zucker et al 2007Structural Interpretation:
Please click HERE for in-depth variant analysis. c.227G>T
p.Cys76Phe (Legacy AA No. 58)
Variant Type:
Point
Domain:
Apple 1
Codon Change:
G>T
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Mitchell et al 2006Structural Interpretation:
Please click HERE for in-depth variant analysis. c.259C>A
p.Pro87Thr (Legacy AA No. 69)
Variant Type:
Point
Domain:
Apple 1
Codon Change:
C>A
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
I
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Quelin et al 2005Structural Interpretation:
Please click HERE for in-depth variant analysis. c.290G>C
p.Gly97Ala (Legacy AA No. 79)
Variant Type:
Point
Domain:
Apple 1
Codon Change:
G>C
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
I
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Gly97Ala occurs in the C terminal portion of Ap1 domain, which contains the binding sites for HK, thrombin and prothrombin. Alanine at position 97 is predicted to introduce two novel hydrogen bonds with the nearby Phe59, probably affecting the local conformation of the protein. Gly97 is strictly conserved in four FXI apple domains of all species and also in kallikrein apple domains. Castaman et al 2008Structural Interpretation:
Please click HERE for in-depth variant analysis. c.296C>A
p.Ser99Tyr (Legacy AA No. 81)
Variant Type:
Point
Domain:
Apple 1
Codon Change:
C>A
Variant Effect:
Missense
No. of Patients Reported:
2
Phenotype:
U
Allele Count *:
1
Allele Number *:
251272
Allele Frequency *:
0.000004
Variant Comments & Reference:
Ser99Tyr is fully buried at the end of β-strand G in the Ap1 domain, thus the replacement with a bulky Tyr residue is expected to disrupt the protein folding. Saunders et al 2009Structural Interpretation:
Please click HERE for in-depth variant analysis. c.302A>G
p.Lys101Arg (Legacy AA No. 83)
Variant Type:
Point
Domain:
Apple 1
Codon Change:
A>G
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
3
Allele Number *:
251288
Allele Frequency *:
0.000012
Variant Comments & Reference:
Lys101 is a highly conserved residue positioned near Cys103, which is involved in a disulphide linkage. Although this substitution involves amino acids with similar physical and chemical properties, the change may affect local conformation. Hill et al 2005Structural Interpretation:
Please click HERE for in-depth variant analysis. c.316C>T
p.Gln106* (Legacy AA No. 88)
Variant Type:
Point
Domain:
Linker
Codon Change:
C>T
Variant Effect:
Nonsense
No. of Patients Reported:
5
Phenotype:
I
Allele Count *:
1
Allele Number *:
251302
Allele Frequency *:
0.000004
Variant Comments & Reference:
Quelin et al 2004Structural Interpretation:
Structural analysis cannot be performed on this (Point | Nonsense) variant. c.325G>A
p.Ala109Thr (Legacy AA No. 91)
Variant Type:
Point
Domain:
Linker
Codon Change:
G>A
Variant Effect:
Missense
No. of Patients Reported:
11
Phenotype:
I
Allele Count *:
6
Allele Number *:
282654
Allele Frequency *:
0.000021
Variant Comments & Reference:
Being located at the last position of exon 4, within a native donor splice site, this mutation may affect the correct splicing of the F11 mRNA. RT-PCR assays performed on total RNA demonstrated that Ala91Thr abolishes the native donor splice site causing the skipping of the affected exon, eventually resulting in a frameshift followed by a premature termination codon. This mutation is predicted to lead to the synthesis of truncated FXI proteins. Such proteins might be recognised as abnormal by cellular quality control systems, resulting in intracellular retention and protein degradation. Mitchell et al 2006, Guella et al 2008, Duncan et al 2008, Tiscia et al 2017, Esteban et al 2017, Colakoglu et al 2018Structural Interpretation:
Please click HERE for in-depth variant analysis. c.328T>G
p.Cys110Gly (Legacy AA No. 92)
Variant Type:
Point
Domain:
Apple 2
Codon Change:
T>G
Variant Effect:
Missense
No. of Patients Reported:
2
Phenotype:
I
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
The Cys110Gly mutation results in the loss of the C110-C193 disulfide bond and likely impairs the structure of the Ap2 domain. Thus, this mutation could alter the assembly and secretion of the protein. Quelin et al 2005Structural Interpretation:
Please click HERE for in-depth variant analysis. c.359T>C
p.Met120Thr (Legacy AA No. 102)
Variant Type:
Point
Domain:
Apple 2
Codon Change:
T>C
Variant Effect:
Missense
No. of Patients Reported:
2
Phenotype:
I
Allele Count *:
2
Allele Number *:
251262
Allele Frequency *:
0.000008
Variant Comments & Reference:
Met120 is a buried residue and is conserved across six species and prekallikrein. Replacement of the hydrophobic non-polar Met with a hydrophilic polar Thr is likely to be structurally disruptive. Expression studies indicate it is not expressed and type I. This amino acid substitution does not prevent the formation of dimers, despite the established role of the Ap2 domain in dimer formation. Mitchell et al 2006, Mitchell et al 2007Structural Interpretation:
Please click HERE for in-depth variant analysis. c.365G>A
p.Gly122Asp (Legacy AA No. 104)
Variant Type:
Point
Domain:
Apple 2
Codon Change:
G>A
Variant Effect:
Missense
No. of Patients Reported:
7
Phenotype:
I
Allele Count *:
2
Allele Number *:
282668
Allele Frequency *:
0.000007
Variant Comments & Reference:
Mitchell et al 2003Structural Interpretation:
Please click HERE for in-depth variant analysis. c.400C>T
p.Gln134* (Legacy AA No. 116)
Variant Type:
Point
Domain:
Apple 2
Codon Change:
C>T
Variant Effect:
Nonsense
No. of Patients Reported:
3
Phenotype:
I
Allele Count *:
8
Allele Number *:
282680
Allele Frequency *:
0.000028
Variant Comments & Reference:
Dossenbach-Glaninger & Hopmeier 2006, Castaman et al 2008Structural Interpretation:
Structural analysis cannot be performed on this (Point | Nonsense) variant. c.403G>T
p.Glu135* (Legacy AA No. 117)
Variant Type:
Point
Domain:
Apple 2
Codon Change:
G>T
Variant Effect:
Nonsense
No. of Patients Reported:
61
Phenotype:
I
Allele Count *:
244
Allele Number *:
282694
Allele Frequency *:
0.000863
Variant Comments & Reference:
The Type II mutation (Glu135*) is the most common mutation in Jewish FXI-deficient patients. Saunders et al 2005, Wiewel-Verschueren et al 2017, Tiscia et al 2017, Colakoglu et al 2018Structural Interpretation:
Structural analysis cannot be performed on this (Point | Nonsense) variant. Unspecified (C136R)
p.Cys136Arg (Legacy AA No. 118)
Variant Type:
Point
Domain:
Apple 2
Codon Change:
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Tiscia et al 2017Structural Interpretation:
Please click HERE for in-depth variant analysis. c.408C>A
p.Cys136* (Legacy AA No. 118)
Variant Type:
Point
Domain:
Apple 2
Codon Change:
C>A
Variant Effect:
Nonsense
No. of Patients Reported:
4
Phenotype:
I
Allele Count *:
3
Allele Number *:
251304
Allele Frequency *:
0.000012
Variant Comments & Reference:
Saunders et al 2009Structural Interpretation:
Structural analysis cannot be performed on this (Point | Nonsense) variant. c.419G>A
p.Cys140Tyr (Legacy AA No. 122)
Variant Type:
Point
Domain:
Apple 2
Codon Change:
G>A
Variant Effect:
Missense
No. of Patients Reported:
5
Phenotype:
I
Allele Count *:
2
Allele Number *:
251282
Allele Frequency *:
0.000008
Variant Comments & Reference:
Cys140 is buried in an alpha-helix of Ap2 and is part of the Cys140-Cys146 conserved disulphide bridge that is (along with two other conserved disulphide bridges) responsible for the correct folding of the Ap domains. Therefore, the Cys140Tyr mutation is likely to significantly impair the folding of the Ap2 domain. Expression studies reveal a secretion defect. Quelin et al 2006, Spena et al 2009Structural Interpretation:
Please click HERE for in-depth variant analysis. c.422C>T
p.Thr141Met (Legacy AA No. 123)
Variant Type:
Point
Domain:
Apple 2
Codon Change:
C>T
Variant Effect:
Missense
No. of Patients Reported:
2
Phenotype:
I
Allele Count *:
11
Allele Number *:
282690
Allele Frequency *:
0.000039
Variant Comments & Reference:
The Thr141Met mutation results in the introduction of a hydrophobic sulfur atom (Met) and the loss of hydrophilic hydroxyl group (Thr). Castaman et al 2008Structural Interpretation:
Please click HERE for in-depth variant analysis. c.423G>A
p.Thr141= (Legacy AA No. 123)
Variant Type:
Point
Domain:
Apple 2
Codon Change:
G>A
Variant Effect:
Silent
No. of Patients Reported:
0
Phenotype:
None
Allele Count *:
1
Allele Number *:
251274
Allele Frequency *:
0.000004
Variant Comments & Reference:
Polymorphism. Wiewel-Verschueren et al 2017Structural Interpretation:
Structural analysis cannot be performed on this (Point | Silent) variant. c.428A>C
p.Asp143Ala (Legacy AA No. 125)
Variant Type:
Point
Domain:
Apple 2
Codon Change:
A>C
Variant Effect:
Missense
No. of Patients Reported:
3
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Jerčić et al 2020Structural Interpretation:
Please click HERE for in-depth variant analysis. c.429C>T
p.Asp143= (Legacy AA No. 125)
Variant Type:
Point
Domain:
Apple 2
Codon Change:
C>T
Variant Effect:
Silent
No. of Patients Reported:
0
Phenotype:
None
Allele Count *:
10089
Allele Number *:
282660
Allele Frequency *:
0.035693
Variant Comments & Reference:
Polymorphism. Ventura et al 2000Structural Interpretation:
Structural analysis cannot be performed on this (Point | Silent) variant. c.434A>G
p.His145Arg (Legacy AA No. 127)
Variant Type:
Point
Domain:
Apple 2
Codon Change:
A>G
Variant Effect:
Missense
No. of Patients Reported:
4
Phenotype:
U
Allele Count *:
39
Allele Number *:
282700
Allele Frequency *:
0.000138
Variant Comments & Reference:
The His145Arg mutation occurs adjacent to the Cys140-Cys146 disulphide pairing in Ap2, responsible for creating the substrate binding site in FXIa. Castaman et al 2007 (ISTH Poster Abstract), Castaman et al 2013, Peng et al 2020Structural Interpretation:
Please click HERE for in-depth variant analysis. c.438C>A
p.Cys146* (Legacy AA No. 128)
Variant Type:
Point
Domain:
Apple 2
Codon Change:
C>A
Variant Effect:
Nonsense
No. of Patients Reported:
22
Phenotype:
I
Allele Count *:
10
Allele Number *:
282710
Allele Frequency *:
0.000035
Variant Comments & Reference:
Founder mutation found in English patients. Peretz et al 1997Structural Interpretation:
Structural analysis cannot be performed on this (Point | Nonsense) variant. c.449C>T
p.Thr150Met (Legacy AA No. 132)
Variant Type:
Point
Domain:
Apple 2
Codon Change:
C>T
Variant Effect:
Missense
No. of Patients Reported:
6
Phenotype:
I
Allele Count *:
1
Allele Number *:
251286
Allele Frequency *:
0.000004
Variant Comments & Reference:
Mitchell et al 2006, Saunders et al 2009, Tiscia et al 2017Structural Interpretation:
Please click HERE for in-depth variant analysis. c.452A>C
p.Tyr151Ser (Legacy AA No. 133)
Variant Type:
Point
Domain:
Apple 2
Codon Change:
A>C
Variant Effect:
Missense
No. of Patients Reported:
4
Phenotype:
I
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Transfection experiments with mutant protein confirmed it is expressed at lower levels comparable to normal FXI. Expression studies confirm type I phenotype (CRM-). Bolton-Maggs et al 2003 (Abstract), Mitchell et al 2006Structural Interpretation:
Please click HERE for in-depth variant analysis. c.452A>G
p.Tyr151Cys (Legacy AA No. 133)
Variant Type:
Point
Domain:
Apple 2
Codon Change:
A>G
Variant Effect:
Missense
No. of Patients Reported:
2
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Mitchell et al 2006Structural Interpretation:
Please click HERE for in-depth variant analysis. c.454G>C
p.Ala152Pro (Legacy AA No. 134)
Variant Type:
Point
Domain:
Apple 2
Codon Change:
G>C
Variant Effect:
Missense
No. of Patients Reported:
2
Phenotype:
U
Allele Count *:
1
Allele Number *:
251280
Allele Frequency *:
0.000004
Variant Comments & Reference:
Saunders et al 2009Structural Interpretation:
Please click HERE for in-depth variant analysis. c.484C>T
p.Arg162Cys (Legacy AA No. 144)
Variant Type:
Point
Domain:
Apple 2
Codon Change:
C>T
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
10
Allele Number *:
251330
Allele Frequency *:
0.000040
Variant Comments & Reference:
Dossenbach-Glaninger & Hopmeier 2003, Tiscia et al 2017Structural Interpretation:
Please click HERE for in-depth variant analysis. c.518G>A
p.Gly173Glu (Legacy AA No. 155)
Variant Type:
Point
Domain:
Apple 2
Codon Change:
G>A
Variant Effect:
Missense
No. of Patients Reported:
3
Phenotype:
II
Allele Count *:
1
Allele Number *:
250424
Allele Frequency *:
0.000004
Variant Comments & Reference:
Functional effect; disturbs proposed FXIa binding loop. Alhaq et al 2000, Mitchell et al 2003, O''Connell et al 2005, Saunders et al 2009Structural Interpretation:
Please click HERE for in-depth variant analysis. c.569T>C
p.Leu190Pro (Legacy AA No. 172)
Variant Type:
Point
Domain:
Apple 2
Codon Change:
T>C
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
3
Allele Number *:
249014
Allele Frequency *:
0.000012
Variant Comments & Reference:
Wu et al 2004 (Abstract)Structural Interpretation:
Please click HERE for in-depth variant analysis. c.596C>T
p.Ala199Val (Legacy AA No. 181)
Variant Type:
Point
Domain:
Apple 3
Codon Change:
C>T
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
I
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Ala199 is located next to the conserved Cys200 residue that forms a disulphide bridge with Cys283 connecting the two ends of the Ap3 domain. The Ala199Val substitution interferes with the disulphide bond leading to a misfolded protein. de Raucourt et al 2008Structural Interpretation:
Please click HERE for in-depth variant analysis. c.599G>C
p.Cys200Ser (Legacy AA No. 182)
Variant Type:
Point
Domain:
Apple 3
Codon Change:
G>C
Variant Effect:
Missense
No. of Patients Reported:
2
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Shao et al 2016, Lin et al 2020Structural Interpretation:
Please click HERE for in-depth variant analysis. c.599G>A
p.Cys200Tyr (Legacy AA No. 182)
Variant Type:
Point
Domain:
Apple 3
Codon Change:
G>A
Variant Effect:
Missense
No. of Patients Reported:
2
Phenotype:
U
Allele Count *:
10
Allele Number *:
251312
Allele Frequency *:
0.000040
Variant Comments & Reference:
Bolton-Maggs et al 2003 (Abstract), Duncan et al 2008Structural Interpretation:
Please click HERE for in-depth variant analysis. c.604A>G
p.Arg202Gly (Legacy AA No. 184)
Variant Type:
Point
Domain:
Apple 3
Codon Change:
A>G
Variant Effect:
Missense
No. of Patients Reported:
2
Phenotype:
II
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Transfection experiments of Arg202Gly-FXI show a 70% reduction in FXI activity, suggesting the Arg202Gly mutation might represent a CRM+ defect. Guella et al 2008Structural Interpretation:
Please click HERE for in-depth variant analysis. c.616T>C
p.Pro206Ser (Legacy AA No. 188)
Variant Type:
Point
Domain:
Apple 3
Codon Change:
T>C
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
2
Allele Number *:
251354
Allele Frequency *:
0.000008
Variant Comments & Reference:
Quelin et al 2006Structural Interpretation:
Please click HERE for in-depth variant analysis. c.646G>A
p.Asp216Asn (Legacy AA No. 198)
Variant Type:
Point
Domain:
Apple 3
Codon Change:
G>A
Variant Effect:
Missense
No. of Patients Reported:
0
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Hopmeier et al 2004Structural Interpretation:
Please click HERE for in-depth variant analysis. c.682C>T
p.Arg228* (Legacy AA No. 210)
Variant Type:
Point
Domain:
Apple 3
Codon Change:
C>T
Variant Effect:
Nonsense
No. of Patients Reported:
2
Phenotype:
I
Allele Count *:
4
Allele Number *:
251444
Allele Frequency *:
0.000016
Variant Comments & Reference:
Saunders et al 2005, Zhang et al 2020Structural Interpretation:
Structural analysis cannot be performed on this (Point | Nonsense) variant. c.688T>C
p.Cys230Arg (Legacy AA No. 212)
Variant Type:
Point
Domain:
Apple 3
Codon Change:
T>C
Variant Effect:
Missense
No. of Patients Reported:
4
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Saunders et al 2005, Tiscia et al 2017Structural Interpretation:
Please click HERE for in-depth variant analysis. c.688T>A
p.Cys230Ser (Legacy AA No. 212)
Variant Type:
Point
Domain:
Apple 3
Codon Change:
T>A
Variant Effect:
Missense
No. of Patients Reported:
4
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Cys230Ser in exon 7 suppresses a disulphide bond that maintains the 220-226 loop in the tertiary structure of the third apple domain. It is also possible that free cysteine affects other disulphide bonds such as the 226-255 bond. Aberrant or absent disulphide bonds may disrupt the protein structure leading to impaired secretion or function or increased clearance of misfolded proteins. Another mutation involving Cys230 has been described (Cys230Arg). The Cys230 residue is buried. As a consequence, the size or charge of the substituting Arg residue may contribute to protein structure disruption. In Cys230Ser however, neither size nor charge can account for the structure disruption, rather the suppression of disulphide bridges is likely to be the determinant of altered secretion. Quelin et al 2009Structural Interpretation:
Please click HERE for in-depth variant analysis. c.695A>C
p.His232Pro (Legacy AA No. 214)
Variant Type:
Point
Domain:
Apple 3
Codon Change:
A>C
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Kawankar et al 2016Structural Interpretation:
Please click HERE for in-depth variant analysis. c.716T>C
p.Phe239Ser (Legacy AA No. 221)
Variant Type:
Point
Domain:
Apple 3
Codon Change:
T>C
Variant Effect:
Missense
No. of Patients Reported:
3
Phenotype:
I
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Transient expression experiment revealed secretion of Phe239Ser FXI was significantly reduced compared with that of wild-type FXI. Morishita et al 2003 (Abstract), Okumura et al 2006Structural Interpretation:
Please click HERE for in-depth variant analysis. c.723C>G
p.Phe241Leu (Legacy AA No. 223)
Variant Type:
Point
Domain:
Apple 3
Codon Change:
C>G
Variant Effect:
Missense
No. of Patients Reported:
0
Phenotype:
U
Allele Count *:
19
Allele Number *:
282784
Allele Frequency *:
0.000067
Variant Comments & Reference:
Asselta et al 2017Structural Interpretation:
Please click HERE for in-depth variant analysis. c.728C>T
p.Ser243Phe (Legacy AA No. 225)
Variant Type:
Point
Domain:
Apple 3
Codon Change:
C>T
Variant Effect:
Missense
No. of Patients Reported:
2
Phenotype:
I
Allele Count *:
1
Allele Number *:
31376
Allele Frequency *:
0.000032
Variant Comments & Reference:
In single transient transfections, FXI-Phe243 levels in media are low compared with FXI-Wild-Type (WT). This appears to be due to reduced secretion, as FXI-Phe243 levels in lysates are comparable to FXI-WT. Co-transfection of FXI-WT with FXI-Phe243 results in reduced FXI in media compared with FXI-WT control, with little change in intracellular protein. Western blots of intracellular protein shows that FXI-Phe243 forms intracellular dimers similar to FXI-WT, indicating that poor secretion of this mutant is not due to a failure to form dimers. Kravtsov et al 2005Structural Interpretation:
Please click HERE for in-depth variant analysis. c.730C>T
p.Gln244* (Legacy AA No. 226)
Variant Type:
Point
Domain:
Apple 3
Codon Change:
C>T
Variant Effect:
Nonsense
No. of Patients Reported:
3
Phenotype:
U
Allele Count *:
3
Allele Number *:
251356
Allele Frequency *:
0.000012
Variant Comments & Reference:
Okumura et al 2006Structural Interpretation:
Structural analysis cannot be performed on this (Point | Nonsense) variant. c.731A>G
p.Gln244Arg (Legacy AA No. 226)
Variant Type:
Point
Domain:
Apple 3
Codon Change:
A>G
Variant Effect:
Missense
No. of Patients Reported:
0
Phenotype:
None
Allele Count *:
601
Allele Number *:
282734
Allele Frequency *:
0.002126
Variant Comments & Reference:
This mutation is seen in compound heterzygosity but FXI:C is consistent with a partial deficiency and is likely a polymorphism. Sun et al 2001, O'Connell et al 2005, Wiewel-Verschueren et al 2017Structural Interpretation:
Please click HERE for in-depth variant analysis. c.738G>C
p.Trp246Cys (Legacy AA No. 228)
Variant Type:
Point
Domain:
Apple 3
Codon Change:
G>C
Variant Effect:
Missense
No. of Patients Reported:
8
Phenotype:
I
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Introduction of an additional Cys residue within the third apple domain of FXI may result in mis-pairing of disulphide bridges resulting in an altered unstable conformation of the protein, targeting it for degradation within the cell. Also the mutant protein may be less efficiently secreted from the cell. Alhaq et al 1999Structural Interpretation:
Please click HERE for in-depth variant analysis. c.738G>A
p.Trp246* (Legacy AA No. 228)
Variant Type:
Point
Domain:
Apple 3
Codon Change:
G>A
Variant Effect:
Nonsense
No. of Patients Reported:
19
Phenotype:
U
Allele Count *:
4
Allele Number *:
251324
Allele Frequency *:
0.000016
Variant Comments & Reference:
Saunders et al 2009, Wang et al 2019, Liu et al 2019, Xia et al 2020, Zhang et al 2020Structural Interpretation:
Structural analysis cannot be performed on this (Point | Nonsense) variant. c.751C>T
p.Gln251* (Legacy AA No. 233)
Variant Type:
Point
Domain:
Apple 3
Codon Change:
C>T
Variant Effect:
Nonsense
No. of Patients Reported:
1
Phenotype:
I
Allele Count *:
1
Allele Number *:
251356
Allele Frequency *:
0.000004
Variant Comments & Reference:
Mitchell et al 2006Structural Interpretation:
Structural analysis cannot be performed on this (Point | Nonsense) variant. c.755G>A
p.Arg252Lys (Legacy AA No. 234)
Variant Type:
Point
Domain:
Apple 3
Codon Change:
G>A
Variant Effect:
Missense
No. of Patients Reported:
1
Phenotype:
U
Allele Count *:
-
Allele Number *:
-
Allele Frequency *:
-
Variant Comments & Reference:
Replacement of hydrophilic basic Arg withhydrophilic basic Lys occurs in the last nucleotide of exon 7 and likely disrupts normal mRNA splicing. Duncan et al 2008Structural Interpretation:
Please click HERE for in-depth variant analysis. c.755G>T
p.Arg252Ile (Legacy AA No. 234)
Variant Type:
Point
Domain:
Apple 3
Codon Change:
G>T
Variant Effect:
Missense
No. of Patients Reported:
0
Phenotype:
U
Allele Count *:
1
Allele Number *:
251340
Allele Frequency *:
0.000004
Variant Comments & Reference:
Bolton-Maggs et al 2003 (Abstract)Structural Interpretation:
Please click HERE for in-depth variant analysis. c.756A>T
p.Arg252Ser (Legacy AA No. 234)
Variant Type:
Point
Domain:
Apple 3
Codon Change:
A>T
Variant Effect:
Missense
No. of Patients Reported:
2
Phenotype:
U
Allele Count *:
1
Allele Number *:
251152
Allele Frequency *:
0.000004